|
JULY: WEEK 5
July, 26th
July, 27th
July, 28th
July, 29th
 Gel run for PCR-modified phasins Gel extraction for phasins recorded the following quantifications:
- Pha-10S-1: X ng/ul
- Pha-10S-2: X ng/ul
- Pha-SS-1: X ng/ul
- Pha-SS-2: X ng/ul
Phasins were digested X-S for 3 hours and were quantificated:
- Pha-10S-1 (X-S): X ng/ul
- Pha-10S-2 (X-S): X ng/ul
- Pha-SS-1 (X-S): X ng/ul
- Pha-SS-2 (X-S): X ng/ul
Competentization of MC1061 (again) because we suspect the presence of a contaminant (it grew without reason on Cm plates).
Transformation of new MC1061 competent cells with:
- 1 ul (4ng) of miniprepped ENTERO-pSB4C5 (positive control);
- 1 ul of RING ligation (RING shouldn't be propagated in MC1061);
- 1 ul of MilliQ (negative control).
Transformed cells have been plated on Cm 12,5 ug/ml agar plates and incubated overnight at 37°C.
Tecan Test
July, 30th
We checked the presence of colonies in plates.
 MC1061 transformed with ENTERO-pSB4C5 (positive control) |  MC1061 transformed with RING |  MC1061 transformed with MilliQ (negative control) |
We calculated (thanks Federica) efficiency as #colonies/ug DNA plated
| Strain | Vector | #colonies | Efficiency
|
| MC1061 | ENTERO-pSB4C5 | 3700 | ~10^6
|
| MC1061 | RING | 0 | -
|
| MC1061 | NOTHING | 0 | -
|
As espected RING and NOTHING didn't grow.
MC1061 competent cells don't replicate RING :) !!!
Miniprep of J04450 to take the vector pSB1A3
3-hour digestion (X-S) and gel run to extract the backbone (~XXXX bp).
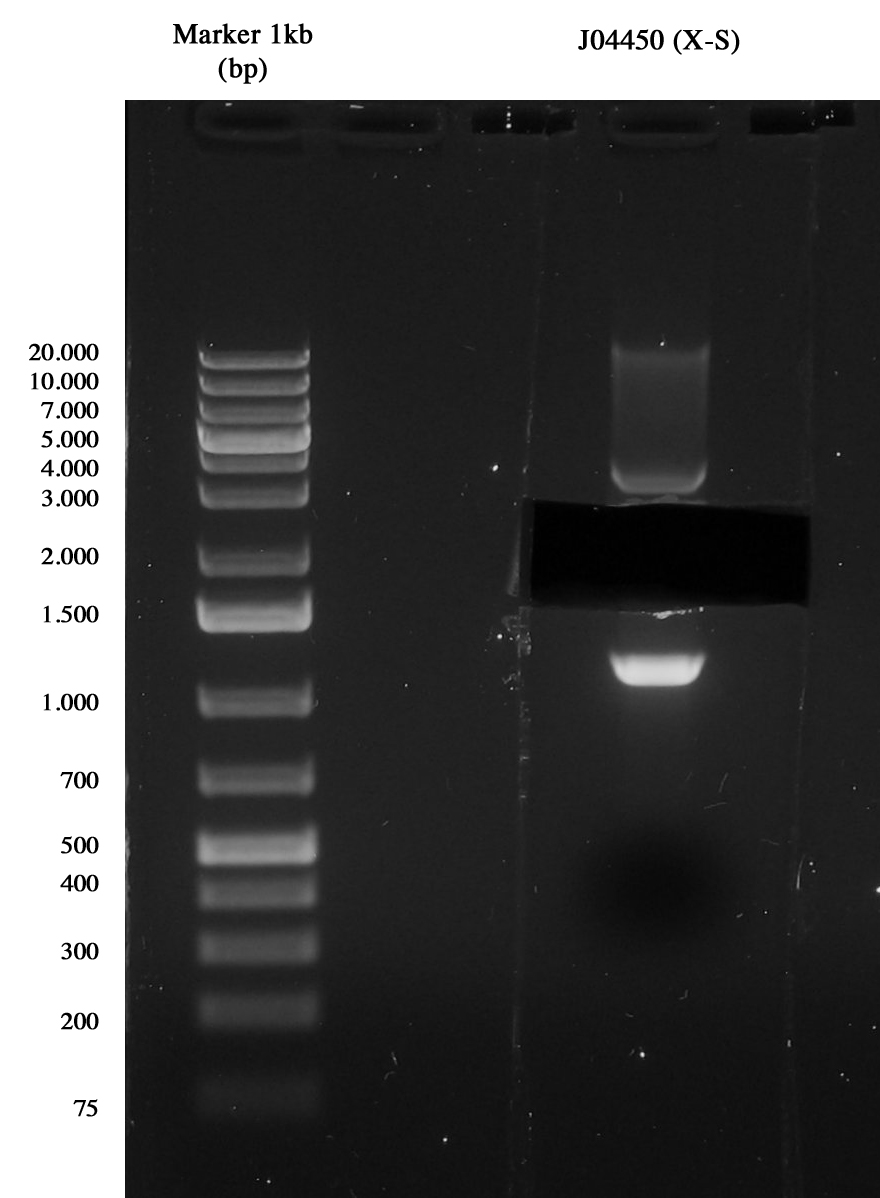 Gel run of J04450 to take pSB1A3 vector Gel extraction was quantificated XX ng/ul.
Ligation of:
- I20: Pha-10S-1 (X-S) + pSB1A3 (X-S)
- I21: Pha-SS-1 (X-S) + pSB1A3 (X-S)
July, 31st
Transformation of I20 and I21 into E. coli DH5-alpha.
|