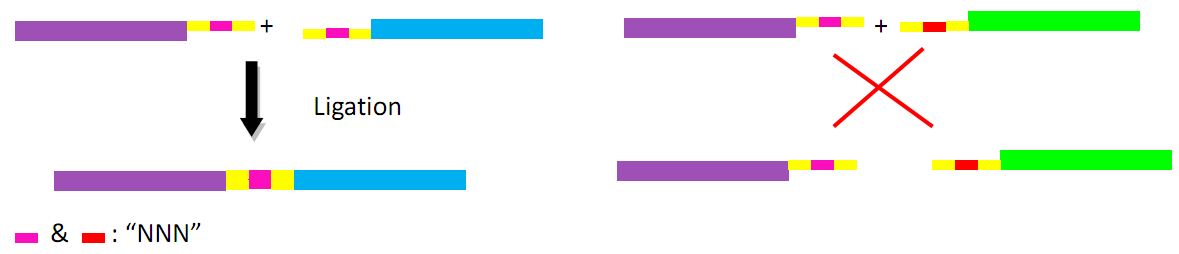Team:Tsinghua/project/outline
From 2010.igem.org

Outline
Module I
Abstract
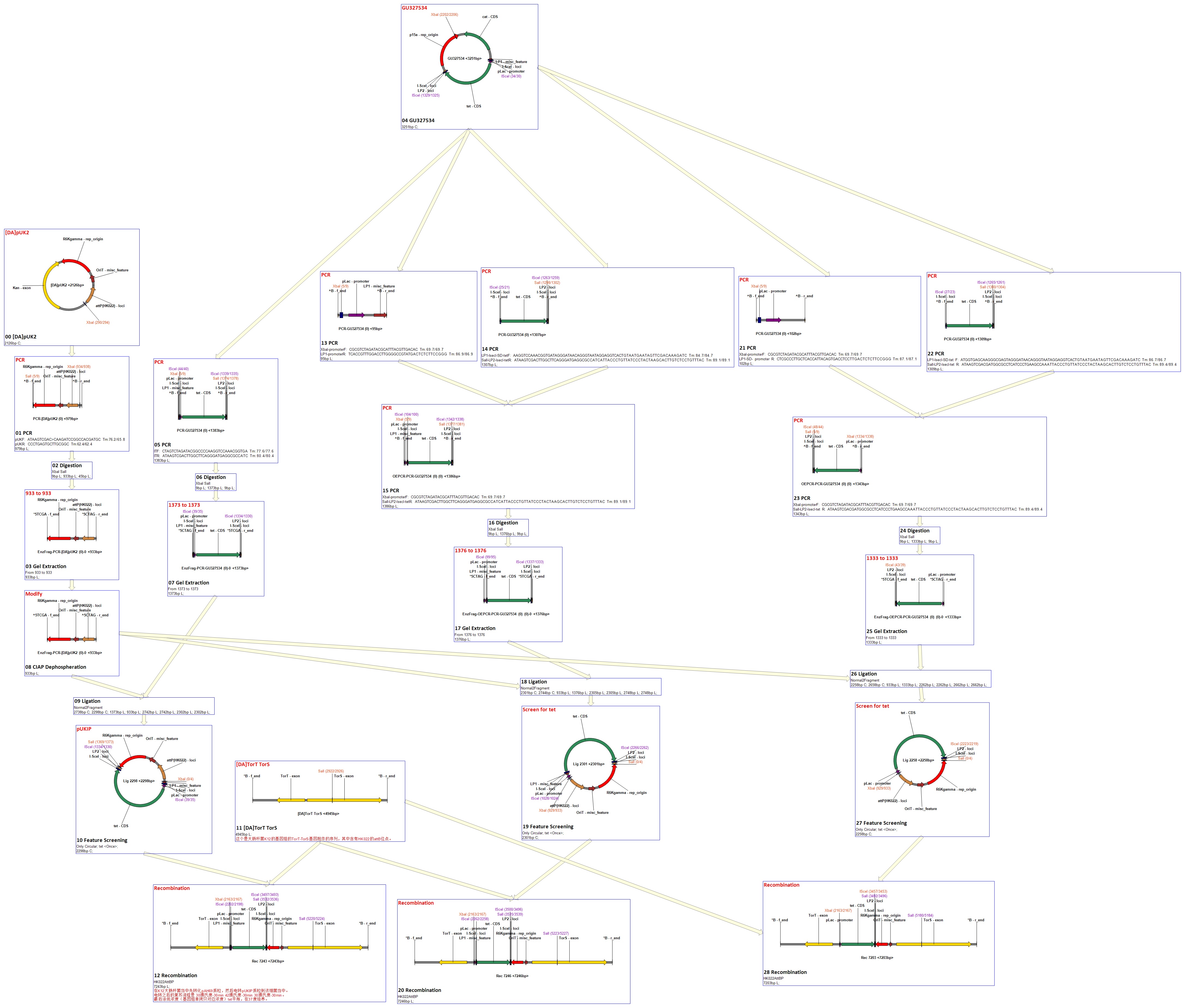
Flow chart
Landing Pad Insertion and Construction
Purpose of this step
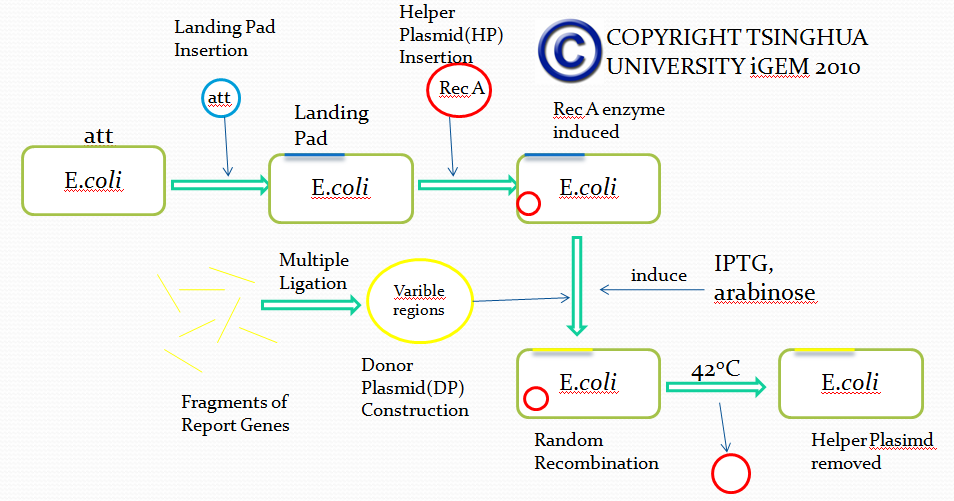
Landing pad insertion is the first step of our two-step recombination system.
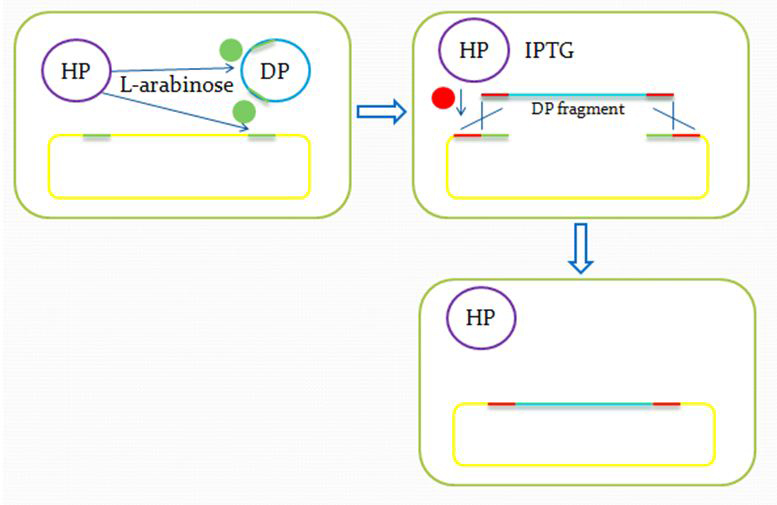
We insert a “landing pad” fragment which includes a promoter (placIQ1) and a tetracycline resistance gene (tetA) flanked by I-SceI recognition sites and 20-bp landing pad regions (LP1 and LP2) into Escherichia coli chromosome via att recombination. Then the helper plasmids encoding I-SceI endonuclease and λ-Red and donor plasmid encoding various antibiotic genes flanked by I-SceI recognition sites and same landing pad regions (LP1 and LP2) are transformed, which will be introduced in detail in next part. I-SceI expression is induced via the addition of L-arabinose. I-SceI recognition sites in the donor plasmid and chromosome are cleaved. Integration of the fragment in donor plasmid is facilitated by IPTG-induced λ-Red expression.
This two-step recombination method allows for the insertion of very large fragments into a specific location in Escherichia coli chromosome and in any orientation.
Construction of landing pad
Landing pad consists of the following parts: a promoter (placIQ1), two landing pad regions, two I-SceI recognition sites and a tetracycline resistance gene (tetA).
First we obtain two target DNAs – promoter and tetA with PCR. Then We use the overlap extension polymerase chain reaction (or OE-PCR) to link two fragments together. Primer 2 and primer 3 have homologous sequences, so one segment can anneal to the other in certain conditions, in other words, the two segments can overlap into one segment after extension. If necessary, operate PCR again with primer 1 and primer 4 to obtain more products.
ATT RECOMBINATION
Our approach is based on genome targeting systems that utilize plasmids carrying a conditional-replication origin and a phage attachment (attP) site (17). We refer to our plasmids as CRIM (conditionalreplication, integration, and modular) plasmids. CRIM plasmids can be integrated into or retrieved from their bacterial attachment (attB) site by supplying phage integrase (Int) without or with excisionase (Xis) in trans.
We got a strain with plasmid pUK2 from LAB. Than we develop a E.coli strain contains the helper plasmid AH69. These two plasmids are shown below. In order to match other parts of our whole project, the modification that kan-exon should be replaced with tet-SDS was necessary. We got tet-SDS from another plasmid named pkts-cs. Then we use PCR to get the two fragments as PT (from pkts-cs) and V (from pUK2). We ligate PT and V after digestion and modification to form a new plasmid. This new plasmid was named as pUKIP and contains a promoter region, a tet-SDS region and phage attachment site (attP). As reported in the paper, there exist one bacterial attachment site (attB) of HK002 in TorT-TorS gene of the chromosomal DNAs of E.coli K12 strain. That is to say, the PT fragment will integrate into the cell genome after the plasmid was transferred in cells.
Helper Plasmid(HP) Insertion
CRIM plasmid integration
Cells carrying a CRIM helper plasmid were grown in 20 ml of LB cultures with ampicillin at 30°C to an optical density of 600 nm of ca. 0.6 and then made electrocompetent. Following electroporation, cells were suspended in LB without ampicillin, incubated at 30°C for 30 min, at 42°C for 30 min and at 30°C for 30 min, and then spread onto selective agar (tet) and incubated at 37°C. Colonies were purified once nonselectively and then tested for antibiotic resistance for stable integration and loss of the helper plasmid and by PCR for copy number.
CRIM plasmid excision
Cells were transformed with the respective Xis/Int CRIM helper plasmid and then spread on ampicillin agar media at 30°C. Colonies were purified once or twice nonselectively on plates that were incubated for 1 h at 42°C and overnight at 37°C. They were then tested for antibiotic sensitivities and by PCR for loss of the integrated plasmid.
Donor Plasmid(DP) Construction
Purpose of this step:
After inserting landing pad and helper plasmid to E.coli, we must construct a series of donor plasmids to demonstrate that this system can truly realize the recombinant process in E.coli, thus we can further use this module to simulate the recombination of antibody gene in mammalian B cells. We not only need to test the efficiency of recombination, but also ensure that genes we get from this recombinant process can be expressed correctly and have their original function. So we intend to construct three plasmids to test the system.
Experiment design and expected results:
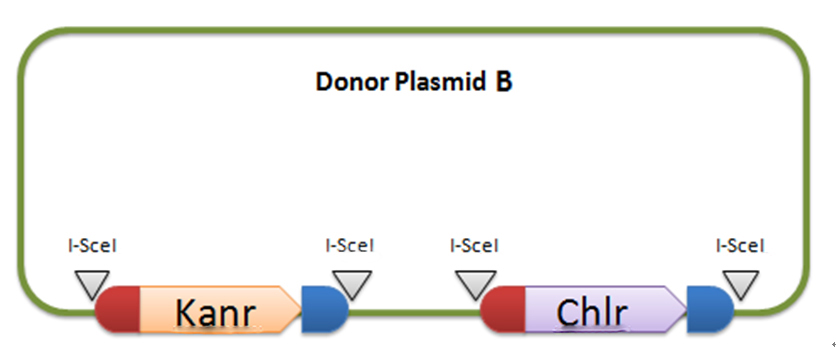
In donor plasmid A, we insert two genes, kanamycin resistant gene (Kanr) and Chloromycetin resistant gene (Chlr). At the 5’ end of these two genes, we add I-scel recognizing sequence (which is represented by the white arrow) and recombination sequence 1 (which is shown in red). At the 3’ end, we add another recombination sequence (which is shown in blue) and also I-scel recognizing sequence. After construction, we will transform this donor plasmid to E.coli with landing pad and helper plasmid. After arabinose and IPTG inducing, the restriction enzyme I-scel will cut down these two genes (containing recombination sequences), which can recombine with the bacterial chromosome. In our expectation, either kanr or chlr will replace the landing pad, resulting in the bacteria resistance to either kanamycin or chloromycetin, but not both. This process will be random, so we can get as many colonies resistant to kanamycin as those resistant to chloromycetin.
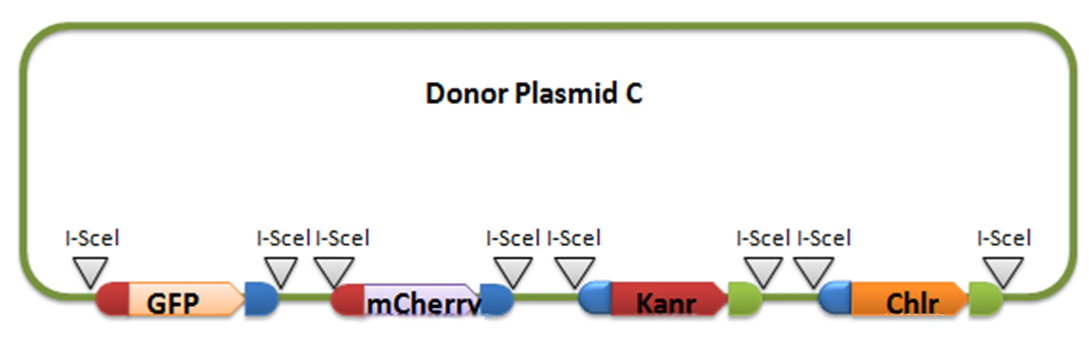
Donor plasmid B includes four genes, GFP, mCherry, Kanr, Chlr, respectively. The same with genes in donor plasmid A, we add recombination sequences and I-scel recognition sequences to the ends of each genes. The recombination sequences of GFP and mCherry are identical, and those of Kanr and Chlr are the same. Note that recombination sequence at 3’ end of GFP (mCherry) and that at 5’ end of Kanr (Chlr) are the same, so we can get a random recombination of two genes, one is a fluorescence gene and the other is resistant gene, creating 2X2=4 different results.
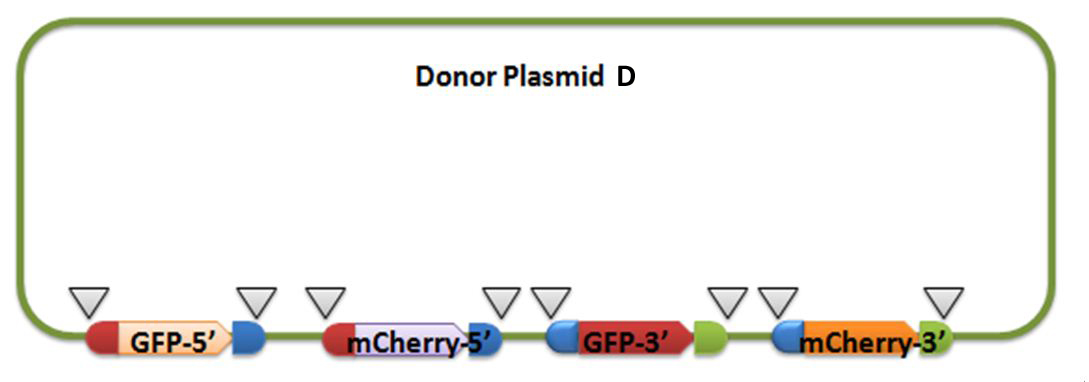
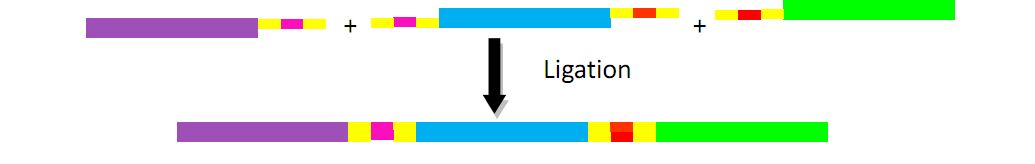
For constructing donor plasmid C, we first cut the GFP and mCherry to 2 fragments respectively. Then we insert these four fragments into the plasmid in the order shown in the above picture. We expect that we can see either green or red fluorescence after transforming and inducing. Through this experiment, we can tell whether or not recombination sequence will affect the normal function of genes, further demonstrate that antibody producing by our system will be effective. On the other hand, we should note that the sequence length is another significant reason of antibody diversity, so the effect of recombination sequence on antibody will much small than that on fluorescence genes.
Strategies
Traditionally, we can construct these three plasmids by inserting genes into the plasmid one by one. However, considering the huge number of antibody fragments, we try our best to seek other strategies to complete the ligation of multiple fragments, which can be done more quickly and efficiently.
Principle
The fragments include different landing pad regions and endoclease recognition site, and are grouped together by sharing the same landing pad region.
Method
In the demand of larger amount of fragments and constructed plasmids, we find two ways to realize such purpose, regarded as the key point of our whole project-guarantee the large variation of antibody.
Removal of Helper Plasmid(HP)
Helper plasmid (HP) includes a temperature sensitive pSC101 replication origin, which maintains the plasmid at low copy number. This plasmid is thus easily removed by growth at 42℃ and screening against spectinomycin resistance. Of more concern is the donor plasmid, which is cured by I-SceI cleavage, and this process is very efficient, with only about 1% of cells retaining the donor plasmid.
Donor Plasmid(DP) Insertion & Recombination Induction
Propose of this step:
After the reform of the E.coli genome and the construction of the donor plasmid, we need to test our module’s function. First we should insert the donor plasmid(DP) into the E.coli, and then induce the recombination by adding IPTG and arabinose. Arabinose active the I-Sel restriction enzyme, then cut DP and genome at the same site. After the digestion, we add IPTG to help the recombination, using the homologous sequence near the cohesive end.
DP Insertion:
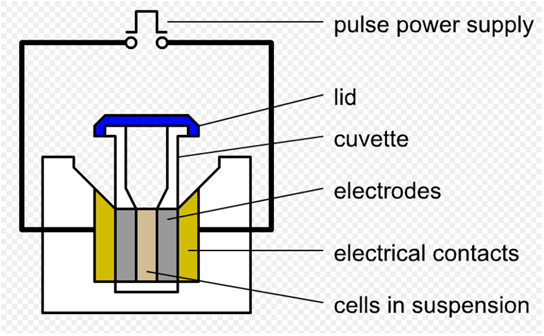
As we said before, we use a pre-altered template to amplify landing pad fragments using the landing pad regions as standardized priming sites. Here we used the conventional electroporation method to transform the DP into E.coli genome, then incubate the plate at 37°overnight. Electroporation is a significant increase in the electrical conductivity and permeability of the cell plasma membrane caused by an externally applied electrical field. It is a dynamic phenomenon that depends on the local transmembrane voltage (we used 1 V) at each point on the cell membrane. If E.coli and DP are mixed together, the plasmids can be transferred into the cell after electroporation. This procedure is highly efficient than chemical transformation. (Partly from wikipedia)
Recombination Induction:
Individual colonies were inoculated into 5 ml of EZ-Rich Defined Medium (RDM; Teknova) +0.5% glycerol, 2mM IPTG, and 0.2% w/v L-arabinose. After growing at 37°C for 1 h in a shaking water bath, we transfer the medium to 30_C shaking water bath for 4 h, then 100 mg/ml spectinomycin was added. At first the I-Sel enzyme is expressed to cut the genome and DP at the same site. And this step is used to constitutive express the Rec A enzyme, thus initiating the recombination by the homologous region. The appropriate antibiotic for the given insertion fragment was then added (25 mg/ml kanamycin, 34 mg/ml chloramphenicol), and the cultures were grown overnight. The next day, 100 ml sample was plated on LB plates with the appropriate antibiotic and grown at 37°C. We test the sample by screening it on LB plates containing 100 mg/ml ampicillin or 10 mg/ml tetracycline to verify the loss of the landing pad and donor plasmid.
Module II
Strategy 1
Bacterial Based Microarray
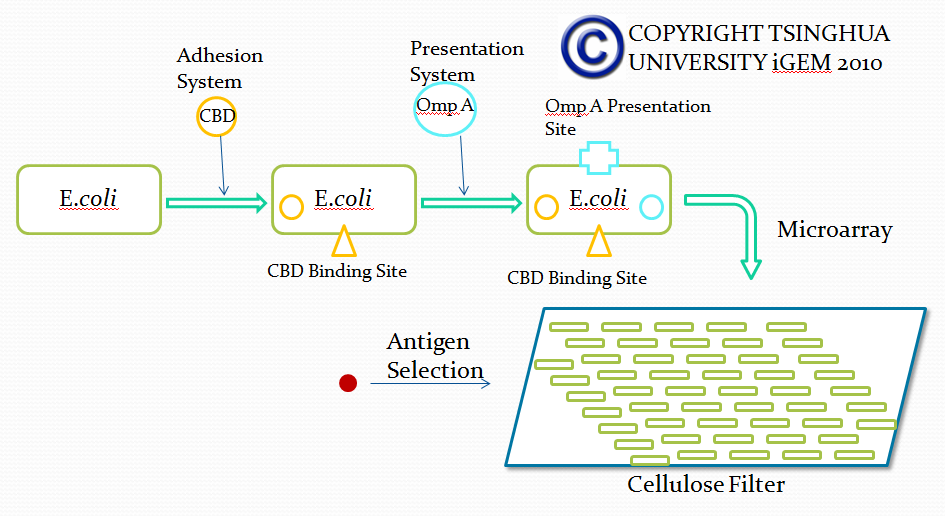
Outline of screening desired antibody by bacterial based microarray
Microarray technique is a kind of pretty mature biomedical technology applied to diverse areas from pharmacology research to clinical use, such as genetics diagnose. Due to the advantage that large amount of various substances can be screened in microarray, microarray can be used for selection of desired antibodies with specific affinity in our project. However, current existing microarray technology rely on specific DNA or protein attached substrate and our project lean on bacteria expressing specific surface protein used for screening for desired antibody. Therefore, traditional protocols and materials used in microarray will be adjusted in our project. We believe that our adjustment will improve the efficiency of antibody screening and help our project achieve its ultimate goal.
Specific description of the details of the experiments
In this part, we want to transform two batches of E coli with vectors carrying specific genes expressing various kinds of antibody achieved by the recombination methods in our project and specific antigen or some other protein used to select antibody from the aforementioned mix of antibodies. By fusion antibody and antigen gene with the genes of membrane integral displaying protein called OmpA, we can manage to display our protein to the surface of the bacteria. Therefore, the selection will process through interaction between antibody and antigen at the surface of the bacteria. Then, by linking the gene of display protein to the gene of one kind of protein called cellulose binding domain which can bind to cellulose, we are able to anchor the bacteria expressing CBD to the surface of the specific microarray coated with cellulose substrate. The whole process is illustrated in the following figure.
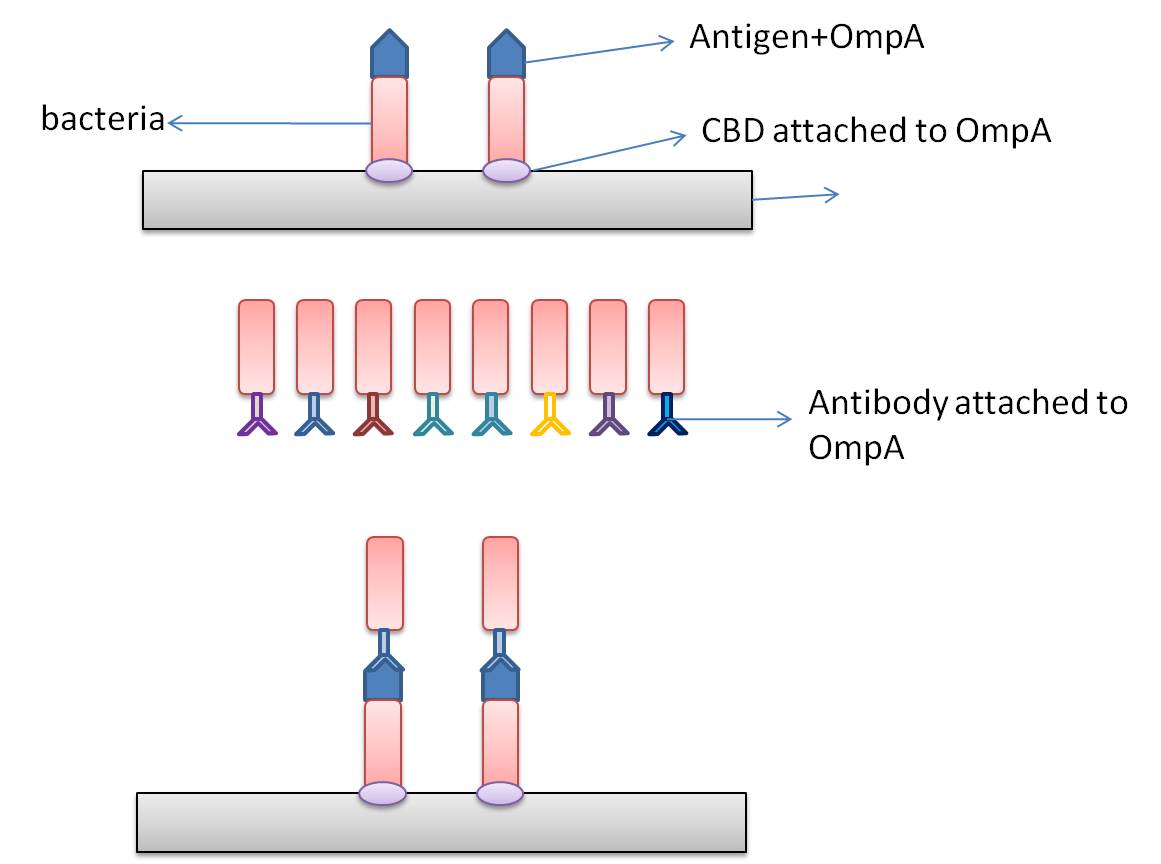
Information concerning the specific proteins
Cellulose binding domain
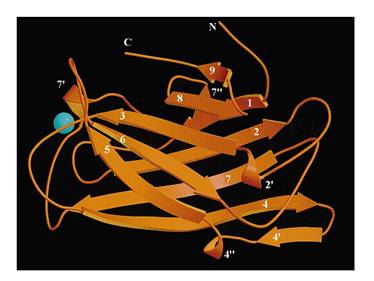
The protein used in this part comes from CipC gene found in Clostridium cellulolyticum. CipC gene encodes a protein called cohesion which participate in degradation of cellulose for the benefits of the bacteria Clostridium cellulolyticum. The protein encoded in this gene contains a specific domain called cellulose binding domain which bind to cellulose with pretty high binding affinity and thus facilitate the degradation of the cellulose. The structure of CBD has been resolved shown in the figure below. Our purpose is to utilize the ability of CBD to bind to cellulose and express CBD in E coli and thus force it to bind to microarray coated with cellulose substrate.
Displaying protein OmpA
At present, two kinds of membrane protein are known, that is, alpha helix protein from cytoplasmic membrane and beta-barrel protein in bacteria outer-membrane, exemplified by porin protein. There are two membrane layers in gram-negative bacteria, separately called inner membrane and outer membrane. OmpA is located in outer membrane and belongs to beta-barrel category. This protein is responsible for normal physiological functions in gene regulation in E coli. Because OmpA is located in outer membrane, intensive investigation has been conducted in OmpA because OmpA plays an important role in signal transduction and much work has focused on the mechanism how OmpA was transported from cytosol out of the membrane. Because of the clarified function of the protein, some bioengineers managed to utilize its function to display protein to achieve certain purposes. Therefore, we try to take advantage of the properties of OmpA to display protein, that is, to display CBD, antibody and antigen. The structure of OmpA has been determined, illustrated in the following figure.
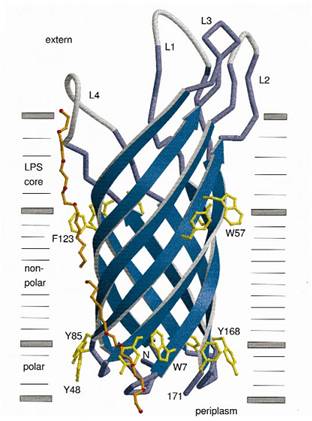
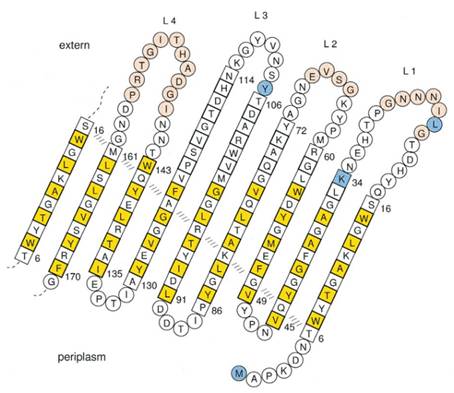
Previous work has demonstrated that 46-159 amino acid is adequate to anchor OmpA to membrane and from the topology figure, we can find out that the fragment of AA 46-159 comprise the transmembrane part of the protein, as the red box indicates. Besides, to ensure the transport of OmpA from cytosol to cell membrane, we need to add a signal peptide to the N terminal of OmpA. Previous work found that one kind of lipoprotein in E coli contains a signal peptide consisting of nine amino acids, which ensures the successful transport of the protein outside the membrane. Mining through the parts provided by iGEM, we found one part which provides exactly the same sequence we wants. This part contains N terminal signal peptide and the sequence AA 46-159 and the linker region downstream of OmpA which ensures the folding of the attached domain, such as CBD. The engineered OmpA is illustrated as following.
The selection of antibodies
Due to commercialization of antibody, we have no access to the cDNA of these antibodies. Therefore, we managed to synthesize the cDNA.
Design of microarray
Microarray is a solid substrate based 2 dimensinoal array, which can be utilized to detect large sum of bio-substance. After decade of developing, various kinds of microarrays have been invented, such as DNA microarray, protein microarray, tissue microarray and so on. Different microarrays can be used to detect different components based on different substrates. Our project belongs to cellular microarray, aiming to select different kinds of antibodies based on the interaction between different proteins displayed on bacteria membrane. This part of our project aims to attach CBD-expressing bacteria to the surface of microarray coated with cellulose. Weizmannn Institute has developed zephyrin-based microarray, in which the microarray plate is dotted with cellulose. Thus, this innovation provides potential for application in our project.
Strategy 2
Transmembrane Signaling Pathway Method
Outline
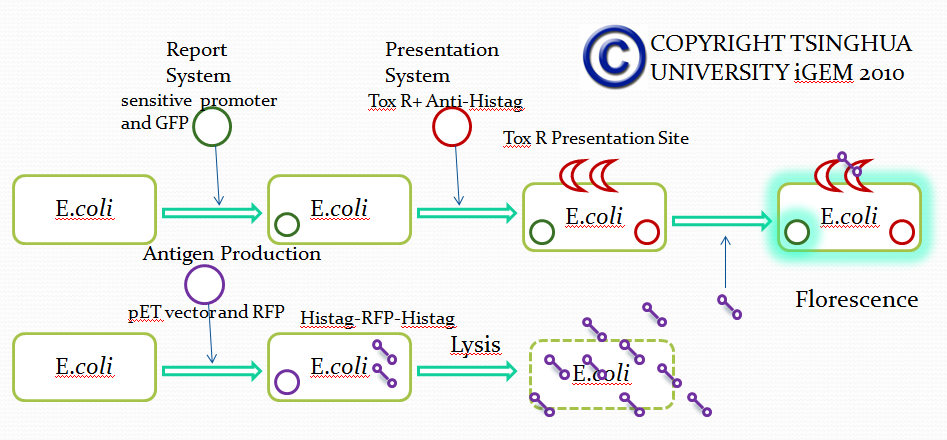
In order to select a specific antibody from a large number of antibodies, we studied the formation of mammalian antibodies. In the process of antibody production in mammals, mediated Annexin antibodies-mediated antibody activation play an important role. This section hopes to use a membrane receptor of E. coli to simulate this process, to achieve the purpose of screening.
To achieve this, a ToxR-based two-hybrid system was introduced into E.coli cells. The Vibrio cholerae transcriptional regulator ToxR is anchored in the cytoplasmic membrane by a single transmembrane segment, its C-terminal domain facing the periplasm. Most of its N-terminal cytoplasmic domain shares sequence similarity with the winged helix–turn–helix (wHTH) motif of OmpR-like transcriptional regulators. The ToxR protein of Vibrio cholerae regulates the expression of several virulence factors that play important roles in the pathogenesis of cholera.
ToxR stimulates transcription from the cholera toxin gene promoter ctx by direct binding to DNA element ITITTGAT present in different isolates of Vcholerae in three to eight tandemly repeated copies upstream of the ctxAB structural genes.
Transcription activation is thought to be initiated by environmental stimuli which cause the periplasmic ToxR domain to form a homodimer. This, in turn, tethers together the two cytoplasmic ToxR domains, which can now bind to the control region of the ctx promoter. A second membraneassociated protein, ToxS (Mr 19 000), is required for maximal activation of the ctx promoter; most plausibly it stabilizes the ToxR dimer by direct contact.
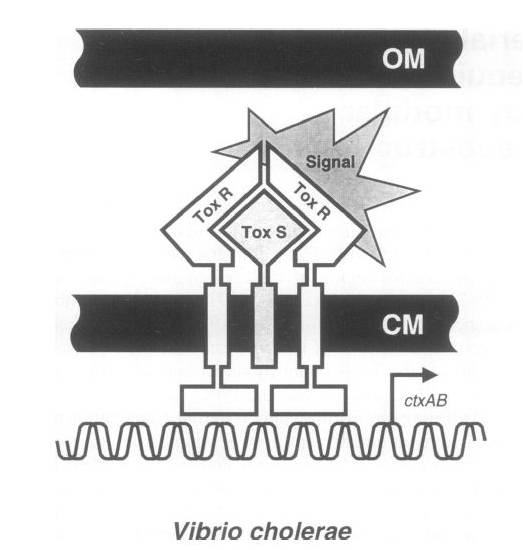 Figure . Original structure of ToxR system in Vibrio cholerae
Figure . Original structure of ToxR system in Vibrio cholerae
Fusion protein was made between the inner- and trans-membrane parts of ToxR and of the recombinant antibody.
When the specific antigens recognized by antibodies, antibodies with the same antigen recognize site will be close to each other through the antigen,inner-membrane part of “Antibody-ToxR” Protein dimerize, ctx promoter active, Resistance gene starts to be transcribed
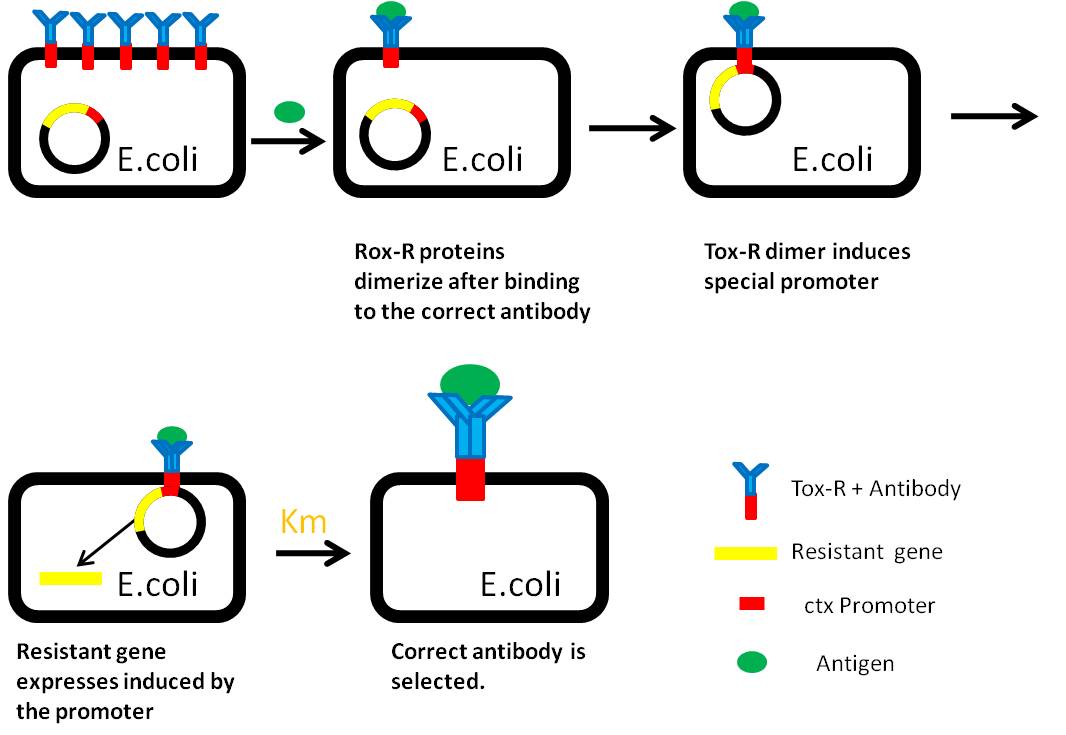
Use “Anti-Histag Antibody” and “Protein with His-tag” as “antibody” and “antigen” to verify the effect of this system.
Antibody: ToxR + Anti-Histag Antibody scFv Light Chain
Plus: ToxR + Anti-Histag Antibody scFv Heavy Chain
Antigen: His tag + Protein A + Histag
Protocol We need to achieve the construction of the selection vector under the control of ctx promoter, then induce to the bacteria. Measure the expression of the two genes and measure the distribution in membrane. Use the exogenous protein carried His-tag as the antigen. Treat with antibiotics. Compare the expression level between the experiment group and the control, then define the effect of our system indirectly.
Reference:A ToxR-based two-hybrid system for the detection of periplasmic and cytoplasmic protein–protein interactions in Escherichia coli: minimal requirements for specific DNA binding and transcriptional activation
Strategy 3
Cooperation with Macquarie Australia
 "
"