Team:TU Munich/Glossary
From 2010.igem.org
(→Linker (modeling)) |
(→Linker (modeling)) |
||
| Line 44: | Line 44: | ||
==Linker (modeling)== | ==Linker (modeling)== | ||
| - | Since Kinefold, the program of choice we used for modeling, can only calculate folding kinetics of individual RNA structures and not the interaction between two structures, which would our case, since it relies on a switch and a corresponding signal, a linker sequence is used to combine both sequences in one. The linker, shown as a row of X can be set to a specific length and does not interact. It mostly serves as a mere placeholder in this model, effects of the linker nevertheless influence the folding time. Since in reality the linker might be thought of as of infinite length, the results shown [https://2010.igem.org/Image:TU_Munich_iGEM2010_sstraub_folding_time.png/ | + | Since Kinefold, the program of choice we used for modeling, can only calculate folding kinetics of individual RNA structures and not the interaction between two structures, which would our case, since it relies on a switch and a corresponding signal, a linker sequence is used to combine both sequences in one. The linker, shown as a row of X can be set to a specific length and does not interact. It mostly serves as a mere placeholder in this model, effects of the linker nevertheless influence the folding time. Since in reality the linker might be thought of as of infinite length, the results shown [https://2010.igem.org/Image:TU_Munich_iGEM2010_sstraub_folding_time.png/ in example of the His-terminator] [https://2010.igem.org/Image:TU_Munich_iGEM2010_sstraub_energy.png/ in the modeling section] considering varying linker lengths actually supports our model: long linkers level the influence of the signal length in time while the minimal free-energy is hardly influenced by the linker length. |
==Logical circuit== | ==Logical circuit== | ||
Revision as of 14:12, 24 October 2010
|
||||||||
|
|
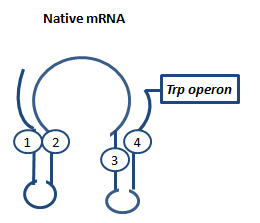
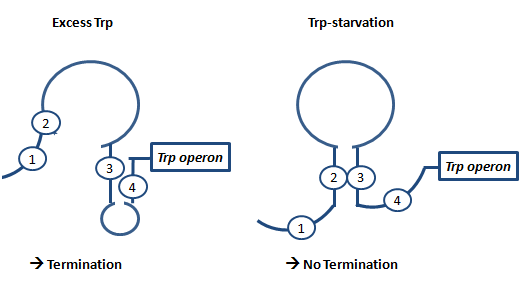
AntiterminationAptamerAptamers in our case are defined as artificially designed single stranded nucleotides designated to specifically bind ligands. Large combinatorial libraries of RNA or DNA molecules are used to isolate and evolve aptamers which achieve surprisingly high affinity and specifity. Aptamers can be used for a broad range of applications: While in our case the malachitegreen-binding aptamer is only used for detection of RNA synthesis in response to an antitermination signal, other approaches use aptamers for regulating gene expression and - together with ribozymes - to design logical gates. By introducing the malachite-green binding aptamer to the Parts Registry we provide a nice tool to evaluate transcription both in vitro and in vivo. Malachite-green binding aptamers were already successfully used for cell cycle control in yeast and aptamers in general were lately tested in mammalian cells in vivo. ZITATE :) AttenuationAttenuation is a very smart way of gene regulation which is known from bacterial cells using two alternatice hairpin structures. For example, E. coli only needs very little amounts of Tryptophan in its metabolism, so the amino-acyl-Synthetase for Tryptophan is only rarely synthesized. So the trp-operon contains an attenuator before the actual enzymes. If Tryptophan is absent, the rare tRNA loaded with Tryptophan will not be available at once, so the Ribosome is stalled. Sterics do not allow the formation of a certain stemloop with the ribosome attached. If there is Tryptophan available and many tRNATrp float through the cell, the ribosome can just continue, a stem loop is formed and the ribosome falls off: The transcription of the following trp-operon is terminated.
E. coliBild von süßem Kuschelcoli iGEMI gonna end up in Massachusetts... Linker (modeling)Since Kinefold, the program of choice we used for modeling, can only calculate folding kinetics of individual RNA structures and not the interaction between two structures, which would our case, since it relies on a switch and a corresponding signal, a linker sequence is used to combine both sequences in one. The linker, shown as a row of X can be set to a specific length and does not interact. It mostly serves as a mere placeholder in this model, effects of the linker nevertheless influence the folding time. Since in reality the linker might be thought of as of infinite length, the results shown in example of the His-terminator in the modeling section considering varying linker lengths actually supports our model: long linkers level the influence of the signal length in time while the minimal free-energy is hardly influenced by the linker length. Logical circuitLogical operatorsRecognition Site (biLOGICS)RiboswitchWhile artificial aptamers are designed to specifally bind their ligands, riboswitches are developed by nature for gene regulation in prokaryotes, with some also found in eukaryotes. Riboswitches are part of mRNAs they control, located in the non-coding parts. They bind to specific metabolites and control gene expression by changing the three-dimensional RNA structures using effects like transcription elongation or translation initiation. Riboswitches typically consist of two parts, the aptamer domain and the expression platform. While the aptamer domain recognizes the metabolite specifically, the expression platforms allows transduction from metabolite binding to gene control by influencing the effciency of translation initiation, transcriptional elongation or stabilizing the mRNA in general. ==Signal (bioLOGICS) Specifity Site (bioLOGICS)SwitchSynthetic BiologyTransistorTrigger Unit (bioLOGICS) |
|||||||
 "
"