Team:SDU-Denmark/project-p
From 2010.igem.org
(Difference between revisions)
(→Motility assay) |
|||
| Line 3: | Line 3: | ||
<div id="subnavi"> | <div id="subnavi"> | ||
<div id="parts"> | <div id="parts"> | ||
| - | + | = Our Parts = | |
<groupparts>iGEM010 SDU-Denmark</groupparts> | <groupparts>iGEM010 SDU-Denmark</groupparts> | ||
<br> | <br> | ||
| - | + | = Characterization of parts = | |
----- | ----- | ||
| - | ==== | + | |
| - | < | + | == K343004 == |
| + | The FlhDC operon is the master regulator of flagella synthesis. A more detailed description of the operon can be found [https://2010.igem.org/Team:SDU-Denmark/project-t#Hyperflagellation here] <br> | ||
| + | In our system the purpose of the composite part is to hyper flagellate our cells so that a grater force can be generated in the microtubes. The FlhDC operon is naturally found in the ''E. coli'' strain MG1655 genome. We extracted the operon and inserted a silent mutation (T to C) at position 822 in the operon because without the mutation this site is a Pst1 digestion site and it would therefor constitute problems when assembling the composit part. <br> We have made three FlhDC parts:<br> [http://partsregistry.org/Part:BBa_K343100 K343100] is the coding sequence of the native FlhDC operon with the Pst1 digestion site <br> [http://partsregistry.org/Part:BBa_K343000 K343000] is the coding sequence of the mutated FlhDC operon <br> [http://partsregistry.org/Part:BBa_K343004 K343004] is the composite part containing the TetR repressable promoter (constitutive when no TetR is pressent) + RBS (J13002), the K343000 part and the double terminator (B0015). <br> | ||
| + | The composite part is caracterized firstly by using a motility assay and secondly by measuring plasmid stability and cell growth. <br><br> | ||
| + | |||
| + | ===Motility assay=== | ||
| + | '''1. Motility assay (Experiment 1 with WT and negative control):''' <br> Growth of bacterial culture on semi-solid agar plates <br> | ||
| + | The purpose of this experiment is to see if the cells containing pSB1C3-K343004 move farther than the wild type (MG1655) and the negative control (DH5alpha). This would be an indication of hyperflagellation. We also want to test if it makes a difference in the motility wether the bacteria contain low-medium- or high-copy plasmids. <br><br> | ||
| + | For the motility experiments we added 5ul of an ON culture to petridishes containing motility agar (LB media with 0.3% agar) instead of regular LA (Luria agar). This semi-solid media lets the bacteria swim more easily. <br> | ||
| + | The plates were incubated at 37 degrees celcius for 24 hours.<br><br> | ||
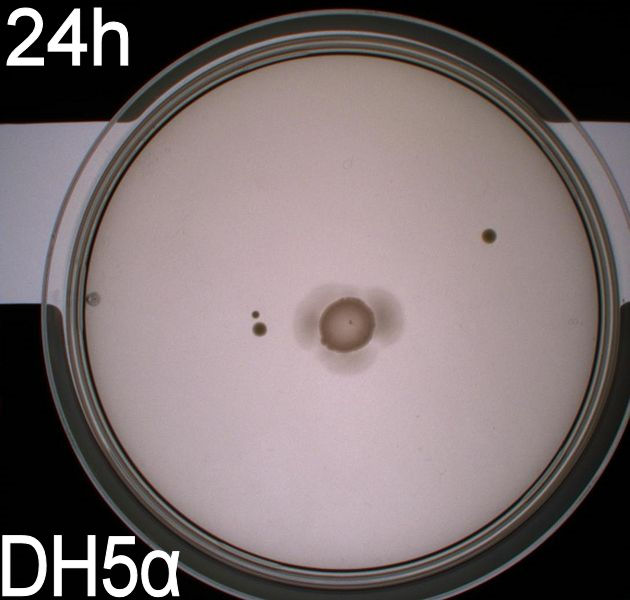
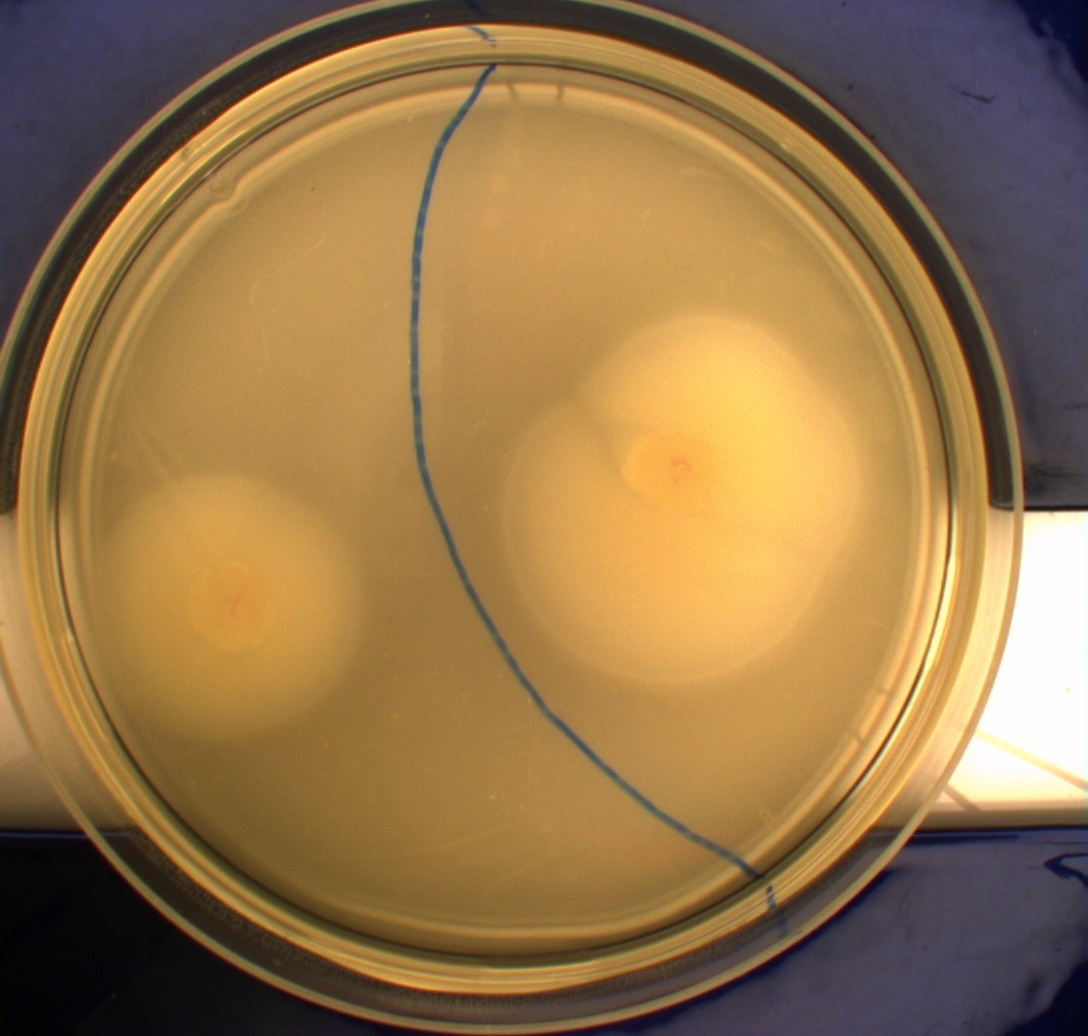
| + | [[Image:Team-SDU-Denmark-Flagellamotility-exp1-DH5a.JPG|250px|DH5alpha]] | ||
| + | [[Image:Team-SDU-Denmark-Flagellamotility-exp1-MG1655.JPG|250px|MG1655]]<br><br> | ||
| + | [[Image:Team-SDU-Denmark-Flagellamotility-exp1-FlhDCmutCP_i_pSB1C3_(LA+chlor).JPG|250px|pSB1C3-FlhDCmutCP(LA+chlor).]] | ||
| + | [[Image:Team-SDU-Denmark-Flagellamotility-exp1-FlhDCmutCP_i_pSB3K3_(LA+kan).JPG|250px|pSB3K3-FlhDCmutCP(LA+Kan)]] | ||
| + | <br><br> | ||
| + | The upper two plates did not contain antibiotics, and therefore contamination colonies are seen.<br> | ||
| + | The upper left picture is of ''E. coli'' strain '''DH5alpha''' that does not express flagella and therefor movement in the media should, as seen on the picture, be minimal. The upper right picture is of the wild type ''E. coli'' strain '''MG1655''', this strain has about 8-10 flagella per cell. These cells are, as seen, expected to move farther than the DH5alpha but not as far as the transformed cells. The lower left picture is of ''E. Coli'' strain '''MG1655 with pSB1C3-K343004''' this shows that these bacteria move farther than the wild type and the negative control. The lover right picture is of ''E. coli'' strain '''MG1655 with pSB3K3-K343004''' these bacteria move farther than the wild type, the negative control and MG1655 with pSB3K3-K343004.<br><br> | ||
| + | |||
| + | From the pictures above we can definately se that the bacteria containing our part is much more motile than the wild type. We assume this is caused by overexpression of the FlhDC master flagella operon which leads to hyperflagellation of the cells. <br> The two buttom pictures show that bacteria with pSB1C3-K343004 have not moved as far as the bacteria containing pSB3K3-K343004. pSB1C3 is a high copy plasmid while pSB3K3 is a low-medium copy plasmid. The promoters in K343004 is a constitutive promoter (tetR repressable promoter). Bacteria containing a high copy plasmid with a constitutive promoter are more metabolically challanged than bacteria containing a low- or medium-copy plasmid with a constitutive promoter because of the higher number of plasmids per the cell. Therefore the high copy plasmid bacteria are less motile than low- or medium-copy plasmid bacteria.<br><br> | ||
| + | ''' 2. Motility assay (doublicate with WT, negative- and positive-control)after 24 hours:''' <br> Growth of bacterial culture on semi-solid agar plates <br><br> | ||
<br> | <br> | ||
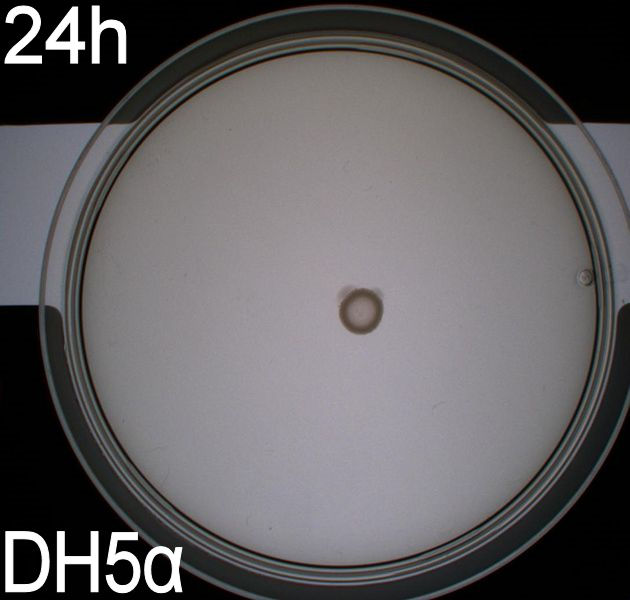
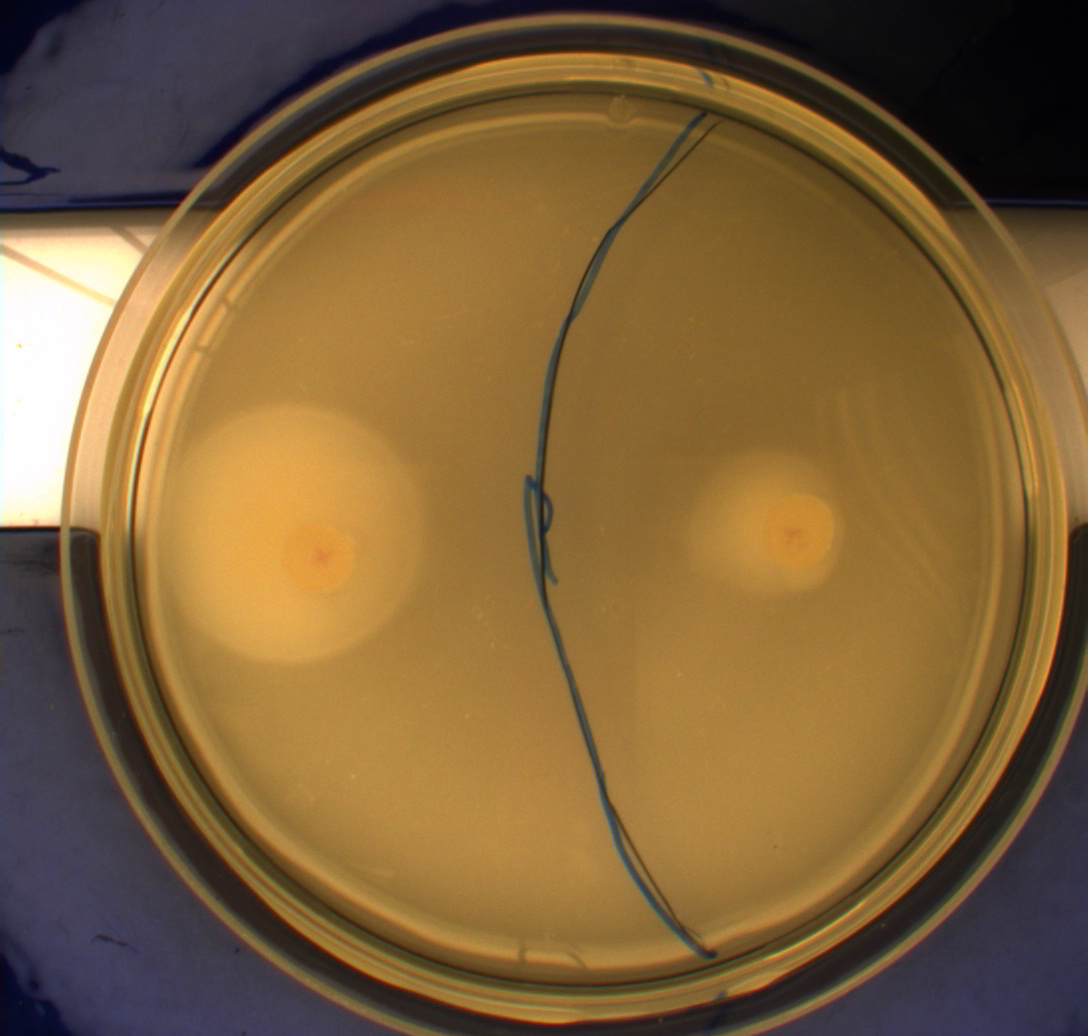
| + | [[Image:Team-SDU-Denmark-Flagellamotility-exp2-DH5a.JPG|250px|DH5alpha]] | ||
| + | [[Image:Team-SDU-Denmark-Flagellamotility-exp2-MG1655.JPG|250px|MG1655]] | ||
| + | [[Image:Team SDU-Denmark exp 2 H10407.JPG|250px|H10407]]<br><br> | ||
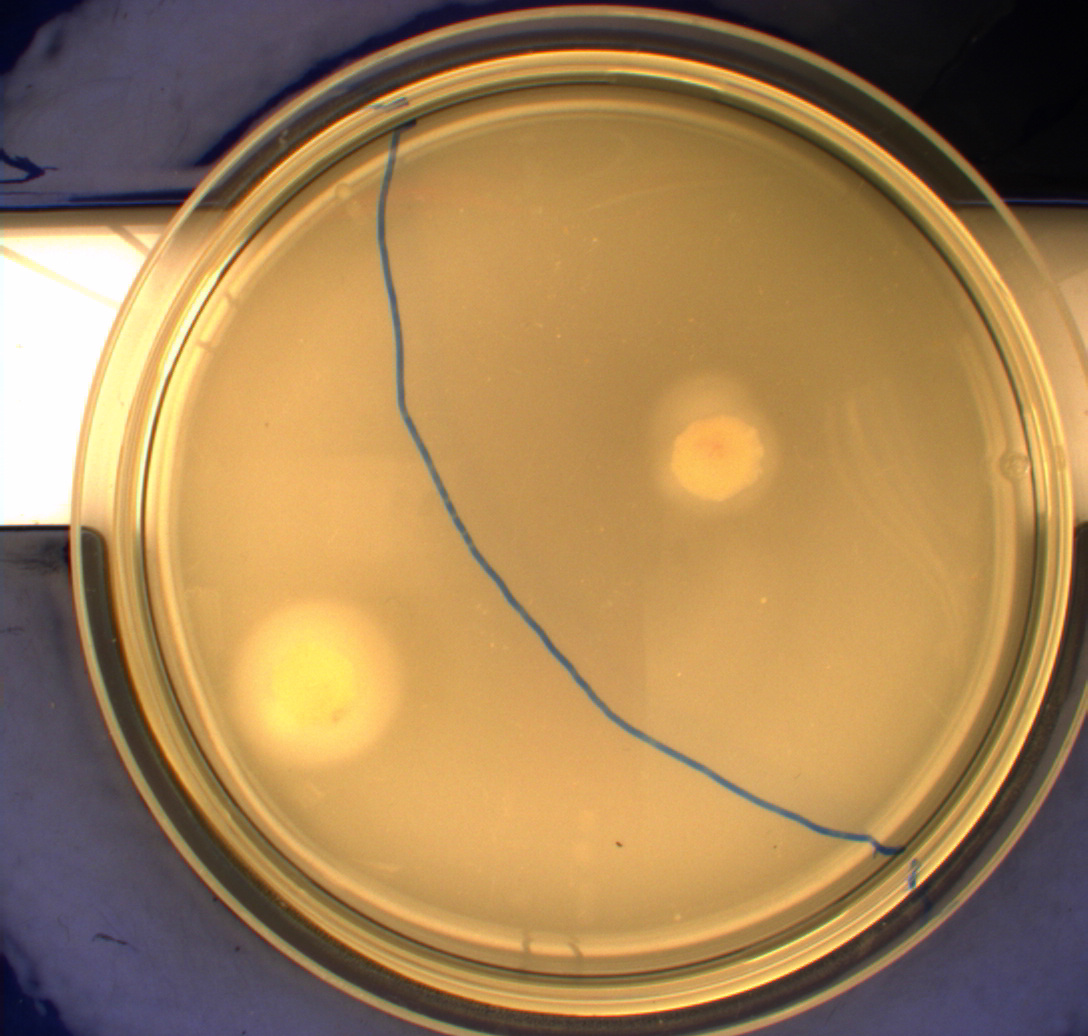
| + | [[Image:Team SDU-Denmark motility exp 2 FlhDCmutCP in pSB1C3.JPG|250px|FlhDCmutCP in pSB1C3]] | ||
| + | [[Image:Team SDU-Denmark motility exp 2 FlhDCmutCP in pSB3K3.JPG|250px|FlhDCmutCP in pSB3K3]]<br><br> | ||
| + | The duplicate of the motility assay shows: <br> | ||
| + | * very little motility of the non-flagellated negative control DH5alpha. Less than in the first assay <br> | ||
| + | * Minimal motility of the wild type MG1655, though they have colonized a bigger area on the plate than DH5aplha. The MG1655 cells also show less motility in this assay than in the first. Also the colony morphorlogy is wvery different in the two assays<br> The only difference between the two assays is that the second assay was plated out and grown on plates in a class II lab because of the positive control. <br> | ||
| + | * Greate motility of the hyper flagellated ''E. coli'' strain H10407 (class II pathogen) positive control <br> | ||
| + | * MG1655 cells with pSB1C3-K343004 have moved farther than both the negative control and the wild type, but not as far as the positive control. On both assays of this transformant a "line" is visable arround the edge of the colony, we think this is an indication that these cells are not moving much further. This transformant show similar motility as in the first assay<br> | ||
| + | * MG1655 cells with pSB3K3-K343004 have moved farthest of all 5 cultures. These cells and the positive control both look as though they are still swimming. This transformant show similar motility as in the first assay <br> | ||
| + | The plates were left in the 37 degrees incubator for another 24 hours to see if our assumptions are correct. <br>''' 3. Motility assay (doublicate with WT, negative- and positive-control)after 48 hours:''' <br> | ||
| + | ''' 4. Stability assay: '''<br> | ||
| + | ''' 5. Growth measurement: ''' <br><br> | ||
| + | |||
| + | == K343007 == | ||
The part K343007 (from now on shortly called PS) is a generator for the SopII-HtrII photosensor from Natronomonas Pharaonis coupled to E.Colis chemotaxis pathway via the Salmonella protein Tar. This part's effect on the system is to make E.Coli phototactic, so that it becomes aware of different light conditions. So for characterization of this part we made examination of motility and motility patterns our first priority and plasmid stability and growth of the cells our second. <br> | The part K343007 (from now on shortly called PS) is a generator for the SopII-HtrII photosensor from Natronomonas Pharaonis coupled to E.Colis chemotaxis pathway via the Salmonella protein Tar. This part's effect on the system is to make E.Coli phototactic, so that it becomes aware of different light conditions. So for characterization of this part we made examination of motility and motility patterns our first priority and plasmid stability and growth of the cells our second. <br> | ||
There is a wide range of motility assays for chemotaxis in bacteria, which meant that we had a broad spectrum of experiments to choose from, which just had to be tweaked for making them suited for the analysis of phototaxis. The two experiments we chose for analysing the effect of this part (PS), were growth of the bacterial cultures in semi-solid agar and computer analysis of swimming motility through video microscopy:<br> | There is a wide range of motility assays for chemotaxis in bacteria, which meant that we had a broad spectrum of experiments to choose from, which just had to be tweaked for making them suited for the analysis of phototaxis. The two experiments we chose for analysing the effect of this part (PS), were growth of the bacterial cultures in semi-solid agar and computer analysis of swimming motility through video microscopy:<br> | ||
| Line 57: | Line 92: | ||
<br></html> | <br></html> | ||
The bacteria that are not moving in the video are assumed to be sticking to the glass or coverslip. | The bacteria that are not moving in the video are assumed to be sticking to the glass or coverslip. | ||
| - | |||
<br> | <br> | ||
| - | + | ''' 5. Stability assay: '''<br> | |
| - | + | ''' 6. Growth measurement: ''' <br><br> | |
| - | + | == K343005 == | |
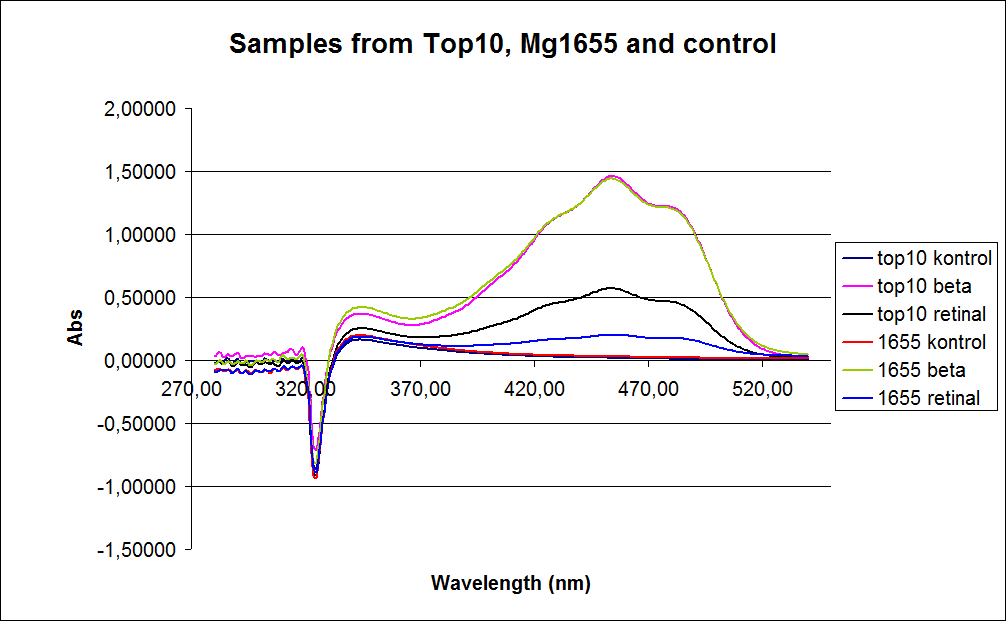
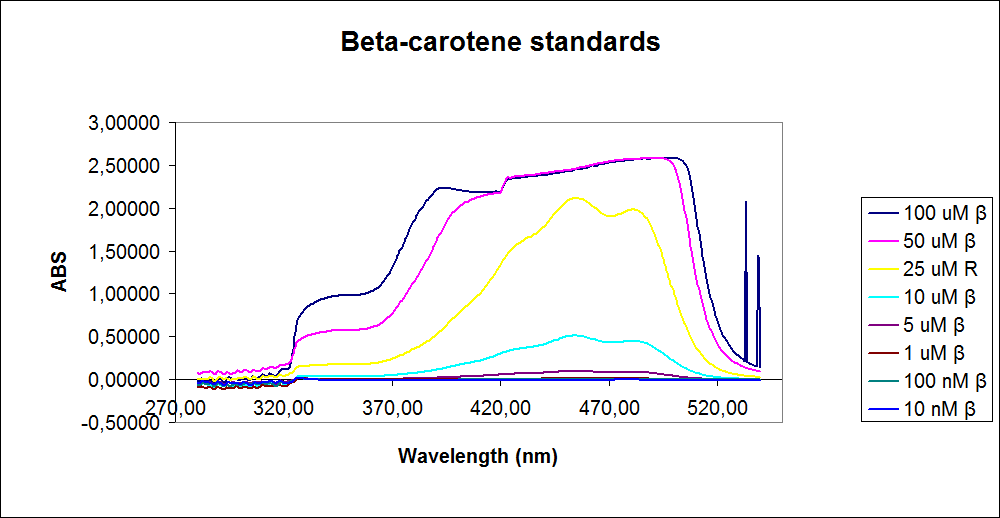
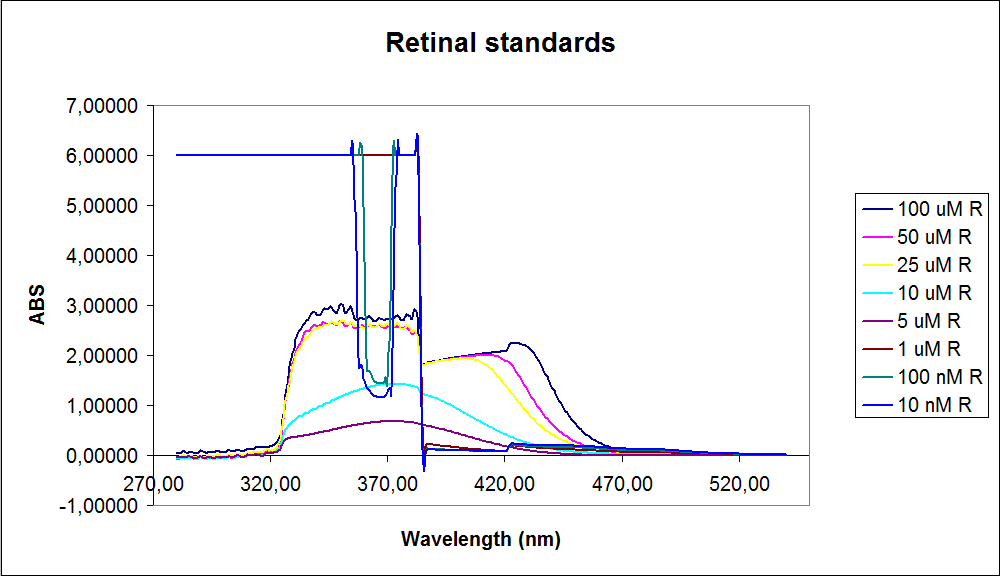
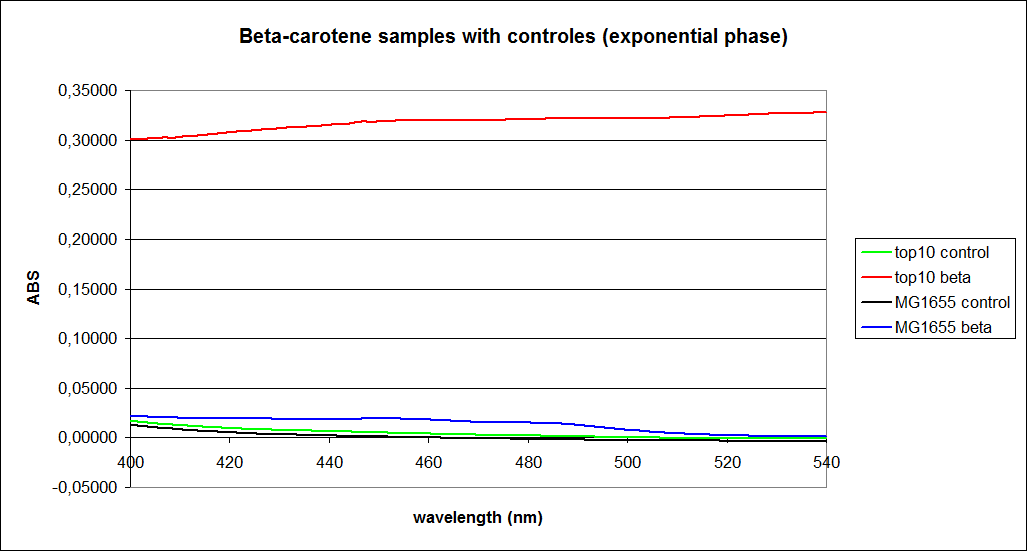
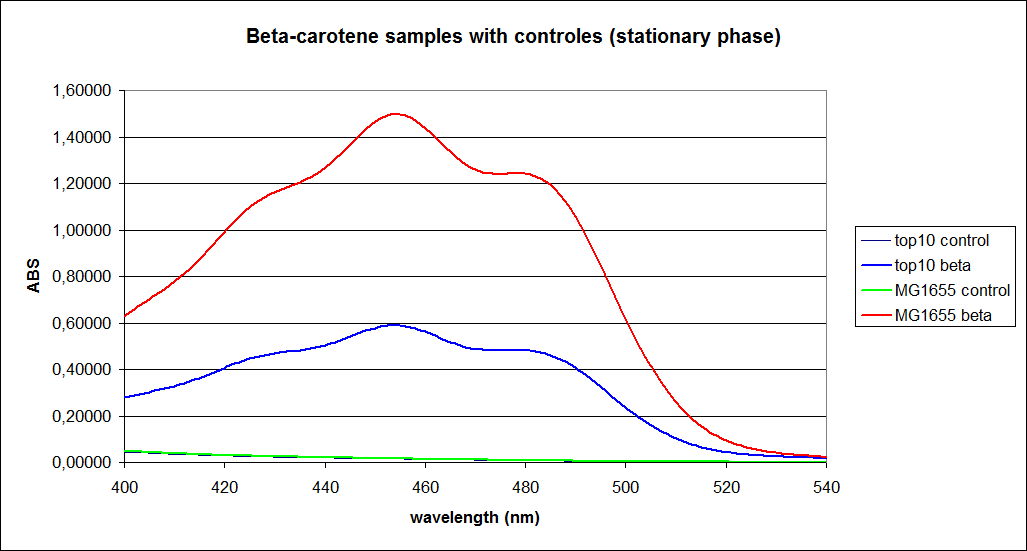
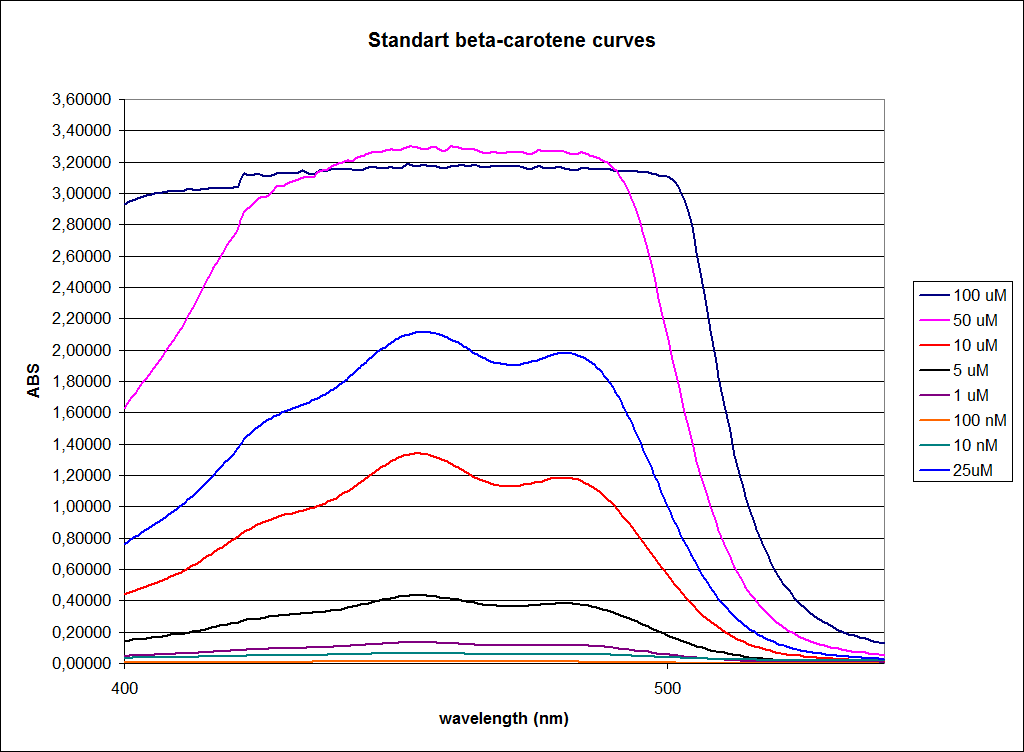
| - | ''' UV-Vis determination of beta-carotene and retinal production '''<br> | + | ''' 1.UV-Vis determination of beta-carotene and retinal production: '''<br> |
In this experiment cells was prepare and harvested according to protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#EX1.1]. This experiment was preformed with six different strain of E. Coli:<br> | In this experiment cells was prepare and harvested according to protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#EX1.1]. This experiment was preformed with six different strain of E. Coli:<br> | ||
wildtype Top10<br> | wildtype Top10<br> | ||
| Line 85: | Line 119: | ||
<br> | <br> | ||
| - | == | + | ==K343006== |
| - | + | ''' 1. HPLC assay: ''' <br> | |
| - | + | ''' 2. Stability assay: '''<br> | |
| - | + | ''' 3. Growth measurement: ''' <br><br> | |
| - | + | ||
| - | + | ||
| - | '''1. | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ''' | + | |
<br> | <br> | ||
| - | + | =Helping another iGEM Team with characterization= | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
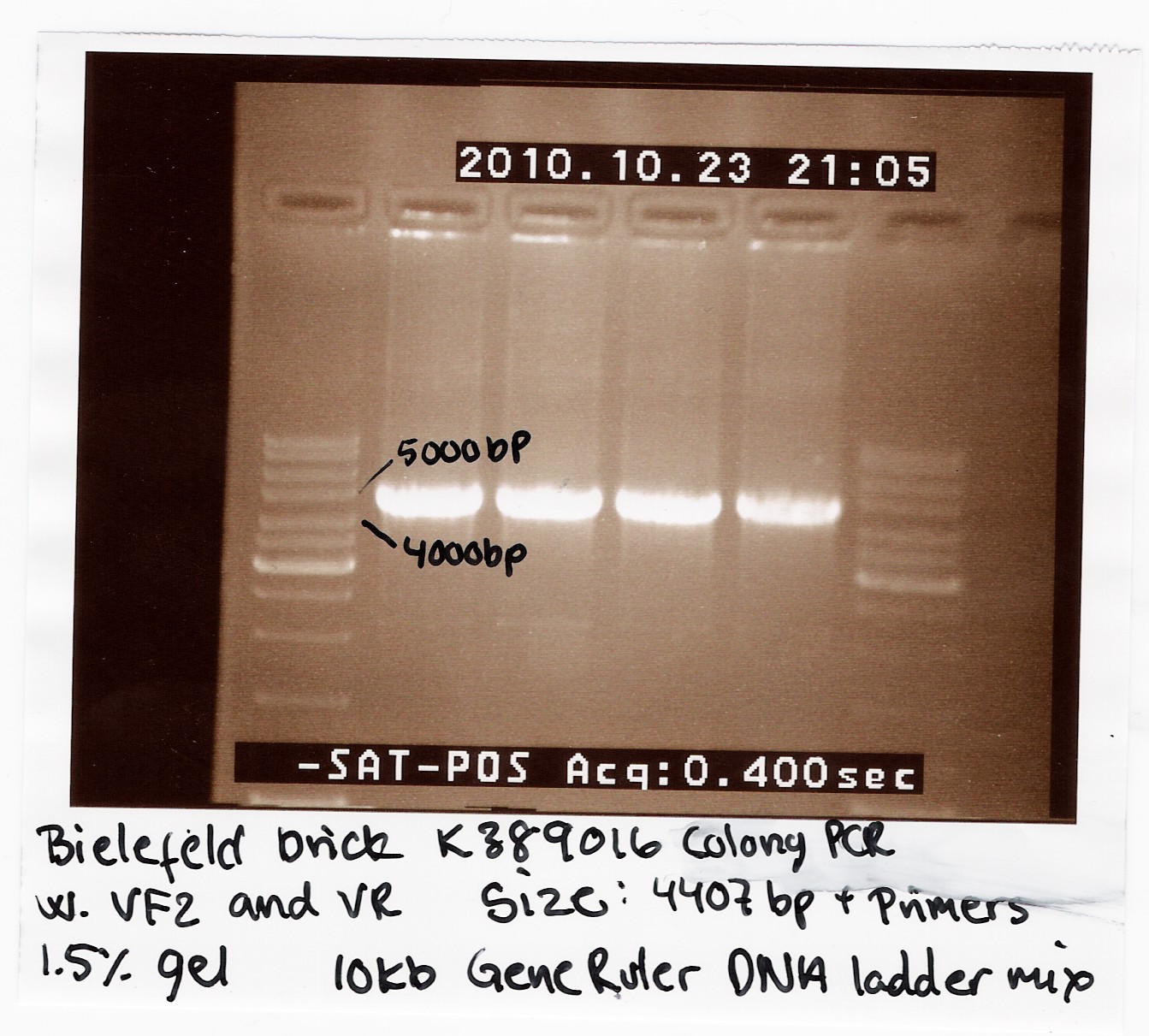
== 2010 [https://2010.igem.org/Team:Bielefeld-Germany Bielefeld] brick K389016 == | == 2010 [https://2010.igem.org/Team:Bielefeld-Germany Bielefeld] brick K389016 == | ||
We recieved a miniprep with [http://partsregistry.org/Part:BBa_K389016 K389016] in pSB1C3 together with a list of experiments and characterization assays the Bielefeld team would like us to carrie out. <br> | We recieved a miniprep with [http://partsregistry.org/Part:BBa_K389016 K389016] in pSB1C3 together with a list of experiments and characterization assays the Bielefeld team would like us to carrie out. <br> | ||
We transformed the plasmid into ''E. coli'' strain MG1655 and ran a colony PCR of four different colonies the following day. The picture below shows that cells in all four colonies contained K389016 in pSB1C3. <br><br> | We transformed the plasmid into ''E. coli'' strain MG1655 and ran a colony PCR of four different colonies the following day. The picture below shows that cells in all four colonies contained K389016 in pSB1C3. <br><br> | ||
| - | [[Image:Team SDU-Denmark Bielefeld Colony PCR-1.jpg | 300px]] | + | [[Image:Team SDU-Denmark Bielefeld Colony PCR-1.jpg | 300px]] <br><br> |
=== Characterization === | === Characterization === | ||
| - | ''' 1. Growth assay ''' <br> | + | ''' 1. Growth assay: ''' <br> |
| + | =Characterizing an exicting biobrick= | ||
== 2009 [https://2009.igem.org/Team:Cambridge Cambridge] brick K274210 == | == 2009 [https://2009.igem.org/Team:Cambridge Cambridge] brick K274210 == | ||
The K274210 brick was cultivated from the 2010 kit plate 3 well N6 in the PSB1A2 plasmid and transformed into E. coli strains TOP10 and MG1655.<br> | The K274210 brick was cultivated from the 2010 kit plate 3 well N6 in the PSB1A2 plasmid and transformed into E. coli strains TOP10 and MG1655.<br> | ||
=== Characterization === | === Characterization === | ||
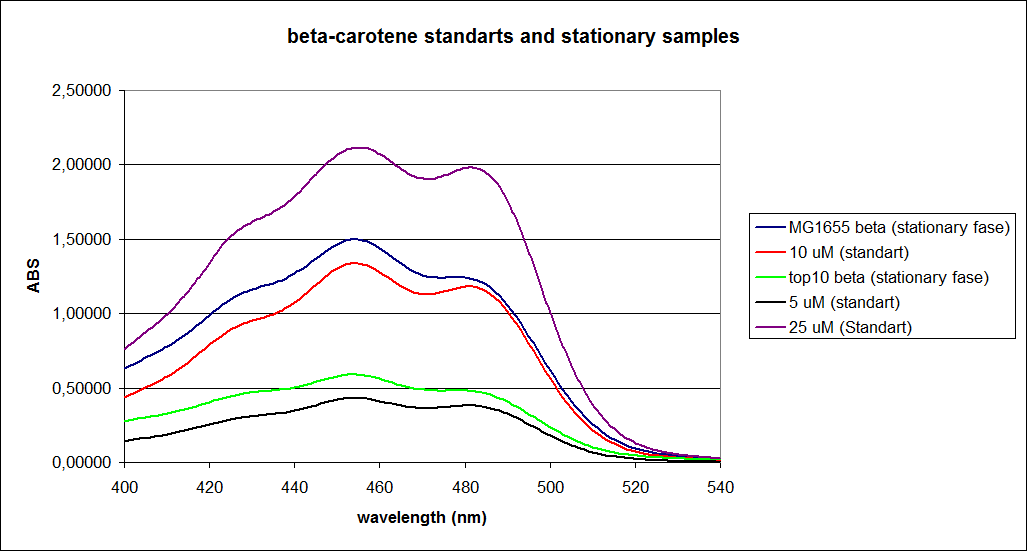
| - | ''' 1. UV-vis determination of beta-carotene production '''<br> | + | ''' 1. UV-vis determination of beta-carotene production: '''<br> |
In this experiment cells were prepared and harvested according to protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#EX1.1]. This experiment was performed with four different strains of ''E. coli'': <br> | In this experiment cells were prepared and harvested according to protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#EX1.1]. This experiment was performed with four different strains of ''E. coli'': <br> | ||
Wildtype Top10 <br> | Wildtype Top10 <br> | ||
Revision as of 23:01, 24 October 2010
 "
"