Team:Mexico-UNAM-CINVESTAV/Notebook/Week One
From 2010.igem.org
(Difference between revisions)
| Line 10: | Line 10: | ||
*''Inmunoresponse using Synthetic Biology'' | *''Inmunoresponse using Synthetic Biology'' | ||
| - | *'' | + | *''Cryobiology applied to plant crops'' |
== '''After a selection process we chose the Criobiology Project''' == | == '''After a selection process we chose the Criobiology Project''' == | ||
Revision as of 20:19, 26 October 2010
As summer project our experimental work began in August, after intensive brainstorming we finnaly decide
between two options.
*Inmunoresponse using Synthetic Biology
*Cryobiology applied to plant crops
After a selection process we chose the Criobiology Project
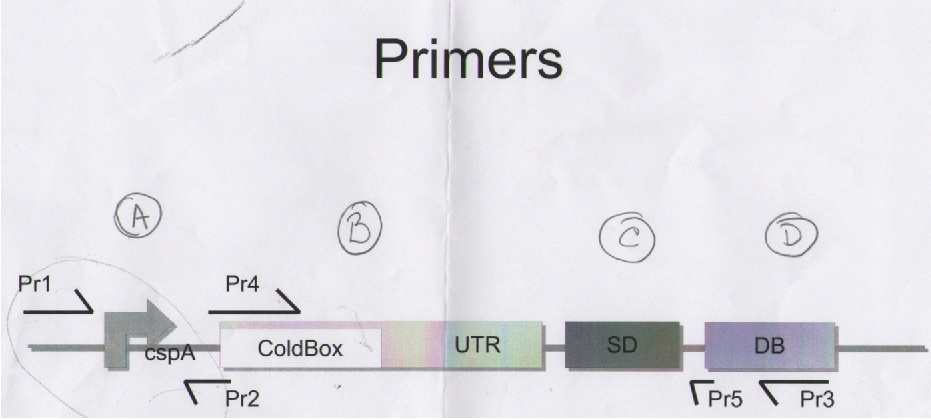
The final design of our expresions moduls is as follow.
Next Notebook paper is our reference for primer's use, ligations and moduls Assembly.
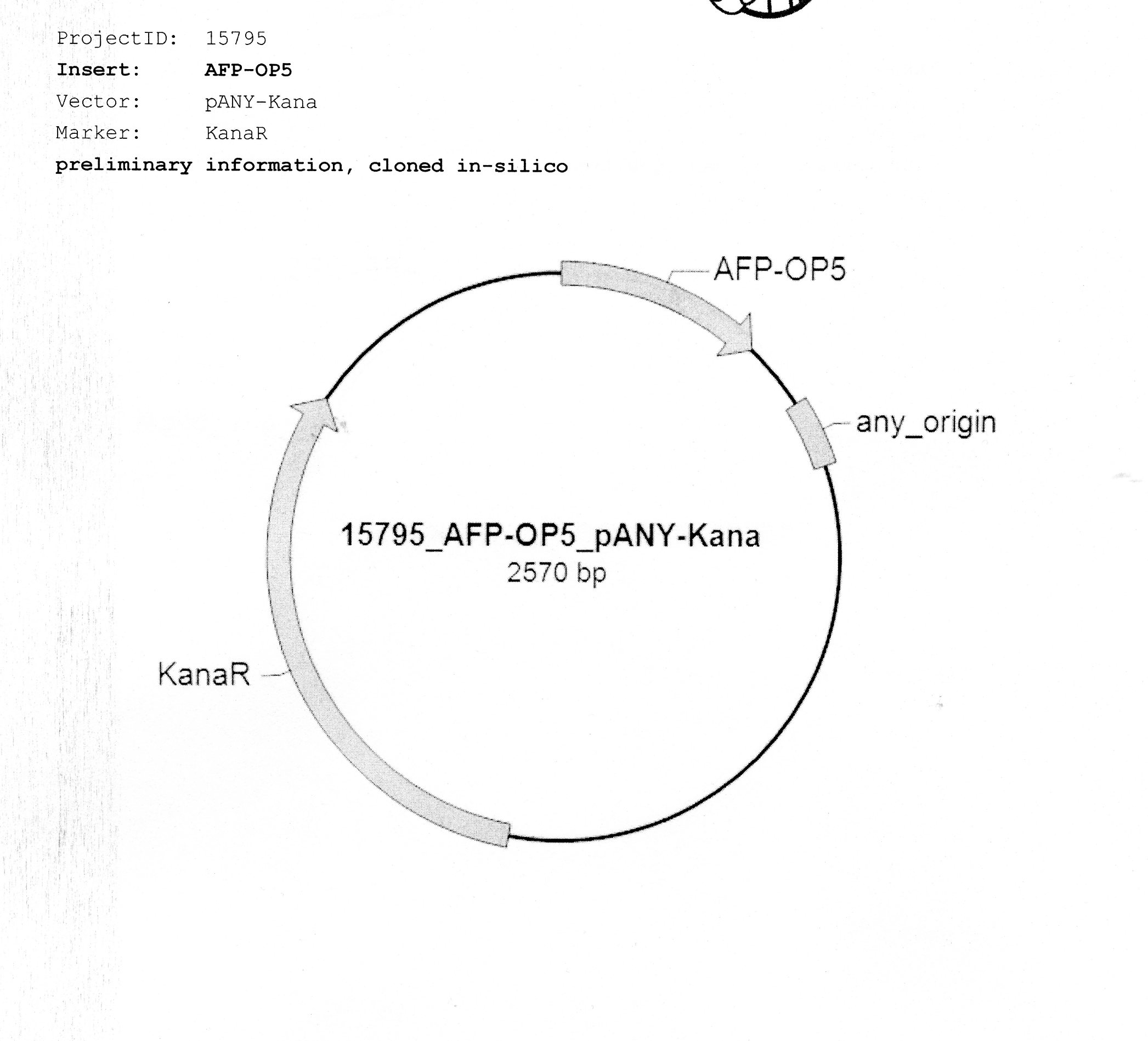
The AFP (Antifreeze protein) was synthesized as follow
Week #1
06th September - 10th September 2010
Monday
We discussed and concluded the Igem’s vector (vial with green cover) is not enough.
First step transform only vector to get enough and begin ligations.
We are going to transform Psb1C3 from plate. For this:
- Prepare a stock of Cloramphenicol, Kanamicyn, Ampicilin solutions.
- 5 LB agar and 35ug/ml cloramphenicol plates made up .
- Complete procedure for making quimio and electro-competent cells.
 "
"