Team:Lethbridge/Results
From 2010.igem.org
Adam.smith4 (Talk | contribs) |
Adam.smith4 (Talk | contribs) (→Results) |
||
| Line 142: | Line 142: | ||
==<b><font color="white">Results</font></b>== | ==<b><font color="white">Results</font></b>== | ||
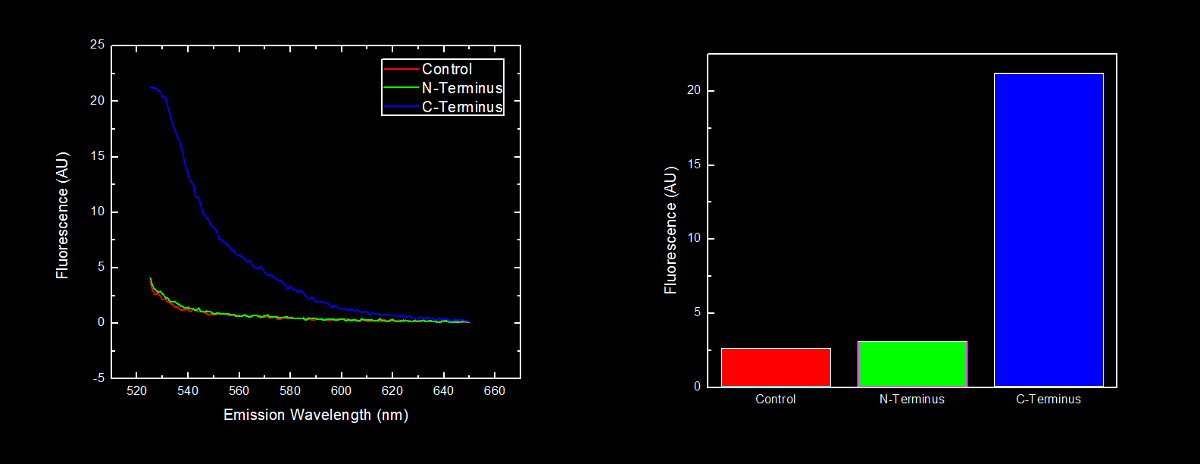
N-terminal tagged YFP did not have substantially more fluorescence than control cells. Cells expressing C-terminal tagged YFP had ten times more fluorescence than control cells and cells expressing N-terminal tagged YFP. | N-terminal tagged YFP did not have substantially more fluorescence than control cells. Cells expressing C-terminal tagged YFP had ten times more fluorescence than control cells and cells expressing N-terminal tagged YFP. | ||
| + | [[image:Lethbridge_NvsC-terminalOligoArgBlackfINAL.png|900px]] | ||
 "
"