Team:Lethbridge/Project
From 2010.igem.org
(Prototype team page) |
(→Project Abstract) |
||
| (55 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | < | + | <div style="background-color:#000000; color:white"> |
| - | + | ||
<html> | <html> | ||
| - | < | + | <br> |
| - | < | + | <table border="0" width="100%" style="background-color:#000000"> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <tr> |
| - | + | <th> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <image src="https://static.igem.org/mediawiki/2010/2/29/UofLteamlogo.jpg" width="200px"/> |
| - | + | </th> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <th> | ||
| + | <image src="https://static.igem.org/mediawiki/2010/9/99/UofLprojectphoto.jpg" height="300px"/> | ||
| + | </th> | ||
| - | + | <th> | |
| - | + | <image src="https://static.igem.org/mediawiki/2010/2/29/UofLteamlogo.jpg" width="200px"/> | |
| + | </th> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | <align="centre"> | ||
| + | <table border="0" width="100%" style="background-color:#000000"> | ||
| + | |||
| + | <tr> | ||
| + | |||
| + | <th> | ||
| + | |||
| + | <a href="https://2010.igem.org/Team:Lethbridge"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/2/22/UofLHome.jpg" width="80"/> | ||
| + | </a> | ||
| + | |||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Team"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/0/0d/UofLTeam.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Project"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/8/8d/UofLProjectbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/7/73/UofLNotebookbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Parts"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/8/84/UofLPartsSubmittedToTheRegistrybutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Modeling"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/e/e1/UofLModelingbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Ethics"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/2/26/UofLEthicsbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Safety"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/0/00/UofLSafetybutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Art"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/0/0a/UofLArt.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/News"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/c/c3/UofLNewsButton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | </table> | ||
| + | </body> | ||
| + | </html> | ||
| + | <hr> | ||
| + | |||
| + | <html> | ||
| + | <center> | ||
| + | <font color="white">If you would like a more detailed look at the different aspects of the project check out these links! | ||
| + | </center> | ||
| + | </html> | ||
| + | |||
| + | <html> | ||
| + | <body> | ||
| + | <center> | ||
| + | <table border="0" width="28%" style="background-color:#000000"> | ||
| + | |||
| + | <tr> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Project/Catechol Degradation"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/d/d0/UofLcatecholbutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Project/Compartamentalization"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/0/0f/UofLcompartmentbutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Project/DNA Degradation"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/0/08/UofLdnabutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Project/Magnetic Nanoparticles"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/e/e3/UofLnanoparticlebutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | </body> | ||
| + | </html> | ||
| + | <hr> | ||
| + | <BLOCKQUOTE> | ||
| + | |||
| + | <!--- The Mission, Experiments ---> | ||
| + | '''Overall project''' | ||
| + | =<font color="white">Project Abstract= | ||
| + | The emerging field of synthetic biology is the combination of engineering approaches with state-of-the-art molecular and cellular biology techniques. It has the potential to change almost every industry. The International Genetically Engineered Machines (iGEM) competition allows undergraduates from diverse backgrounds to explore research in synthetic biology. The University of Lethbridge 2010 iGEM team has created a synthetic-biology-based approach for bioremediation of the tailings ponds. The industrial methods, used to harvest the oil sands, produce contaminated water in the form of tailings ponds that contain many harmful chemicals such as naphthenic acids, catechol and heavy metals. We are targeting catechol for degradation into a common metabolic intermediate that can be fed into the Krebs Cycle by using <i>xylE</i> from <i>Pseudomonas putida</i> that codes for the protein <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Catechol_Degradation"><font color="#00DC00"> catechol-2,3-dioxygenase</font></a></html>. Catechol-2,3-dioxygenase is being targeted into <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Compartamentalization"><font color="#00DC00"> microcompartments</font></a></html>, formed from an engineered <i>Aquifex aeolicus</i> protein, lumazine synthase, to reduce cross-talk and increase concentration. This complex can then be purified and applied to the tailings for catechol degradation. By funnelling other pathways through catechol we can develop efficient methods for the decontamination of the tailings ponds. <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Magnetic_Nanoparticles"><font color="#00DC00"> <i>Mms6</i></font></a></html> from <i>Magnetospirillum magneticum</i> has been shown to remove heavy metals from solution. The <i>Mms6</i> protein will be engineered for secretion from the cell into the tailings for the formation of nanoparticles such as iron and cobalt. Removal of the heavy metals found within the tailing will aid in creating an efficient bioremediation process. | ||
| + | =<font color="white">Oil Sands Initiative Project Proposal= | ||
| - | == | + | ==<font color="white">Reducing the Environmental Footprint of Oil Sands Extraction== |
| + | ===<font color="white">Why are we interested?=== | ||
| + | As a team from Alberta, we know of the economic benefit that the oils sands have brought to the province. Many <html><a href="https://2010.igem.org/Team:Lethbridge/Team"><font color="#00DC00"> members on our team</font></a></html> have friends and family employed around the province in the energy industry, however, we are also aware of the environmental impact of the oils sands, specifically the tailings ponds. We feel it is important not only to clean the tailings ponds, but be able to extract the potential energy of the residue bitumen concurrently. Our team has previously worked on related projects in past years providing us with experience in techniques required to complete this project and potential problems that may arise. Also, being able to work on a project where we will be able to test our bacteria on real life samples gives us a physical goal as opposed to other, more esoteric projects. | ||
| + | ===<font color="white">The Approach=== | ||
| + | The long term goal of our project will be to create a dry powder that will enzymatically degrade tailing ponds with no live bacteria, similar to how the pills taken for lactose intolerance contain enzymes for dealing with lactose. However this will require many intermediate steps. The initial goal of our project will be to create three separate modules for use in a bacteria test chassis. These modules will 1) <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Catechol_Degradation"><font color="#00DC00">degrade target contaminants into useful metabolites</font></a></html>, 2) <html><a href="https://2010.igem.org/Team:Lethbridge/Project/DNA_Degradation"><font color="#00DC00">induce cell death</font></a></html> and 3) <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Compartamentalization"><font color="#00DC00">concentrate the modules in microcompartments</font></a></html>. Upon successful testing of each module, they will be combined into one cell chassis (<i>E. coli</i>). This chassis will then be optimized to live in a tailing pond environment by selective evolution. By combining our modules in microcompartments, we can extract the enzymes required to process tailings ponds and avoid putting live genetic organisms into the wild. | ||
| - | === | + | ===<font color="white">The Modules=== |
| + | <ol> | ||
| + | <li>Bacteria have been found in tailings ponds and even in asphault. They have been shown to be able to degrade many organic compounds in nature. The pathways responsible for degradation of aromatic hydrocarbons produce catechol, a colourless, ortho-substituted benzene with two hydroxyl groups as an intermediate. Our goal is to increase the rate of <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Catechol_Degradation"><font color="#00DC00">catechol degradation</font></a></html> by optimizing the pathway for introduction into the tailings. We will use a pathway which degrades catechol into a semialdehyde which is usable by the bacteria and can be further degraded into usable resources such as ethanol or methanol.</li> | ||
| + | <li>Previous iGEM teams have been able to remotely activate <i>E coli’s</i> <html><a href="https://2010.igem.org/Team:Lethbridge/Project/DNA_Degradation"><font color="#00DC00">DNA degradation pathway</font></a></html> which is the pathway responsible for terminating the cell. This is a very powerful tool, as part of the apoptosis pathway degrades the cell’s DNA. By providing the ability to destroy our custom cell through DNA degradation, we ensure that the cell will not escape into the environment, the DNA will be degraded eliminating the potential for horizontal gene transfer between other species present in the tailing ponds, and the relevant enzymes will not be destroyed.</li> | ||
| + | <li>Our iGEM project last year was the assembly of <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Compartamentalization"><font color="#00DC00">synthetic microcompartments</font></a></html> to isolate pathways from the rest of the cellular environment. These microcompartments will be used to house the catechol degradation pathway so that the pathway can be isolated from the cell and administered to the tailings in a powdered form.</li></ol> | ||
| + | ===<font color="white">Optimization=== | ||
| - | + | Using selective evolution in the presence of catechol we will engineer an <i>E. coli</i> which is able to live in high concentrations of catechol. This will allow for optimization of the catechol degradation pathway using. The next step will be to further use selective evolution with the tailings sample so that the pathway will be optimized for the environmental conditions that the pathway will administered to. | |
| + | ===<font color="white">Our Expectations=== | ||
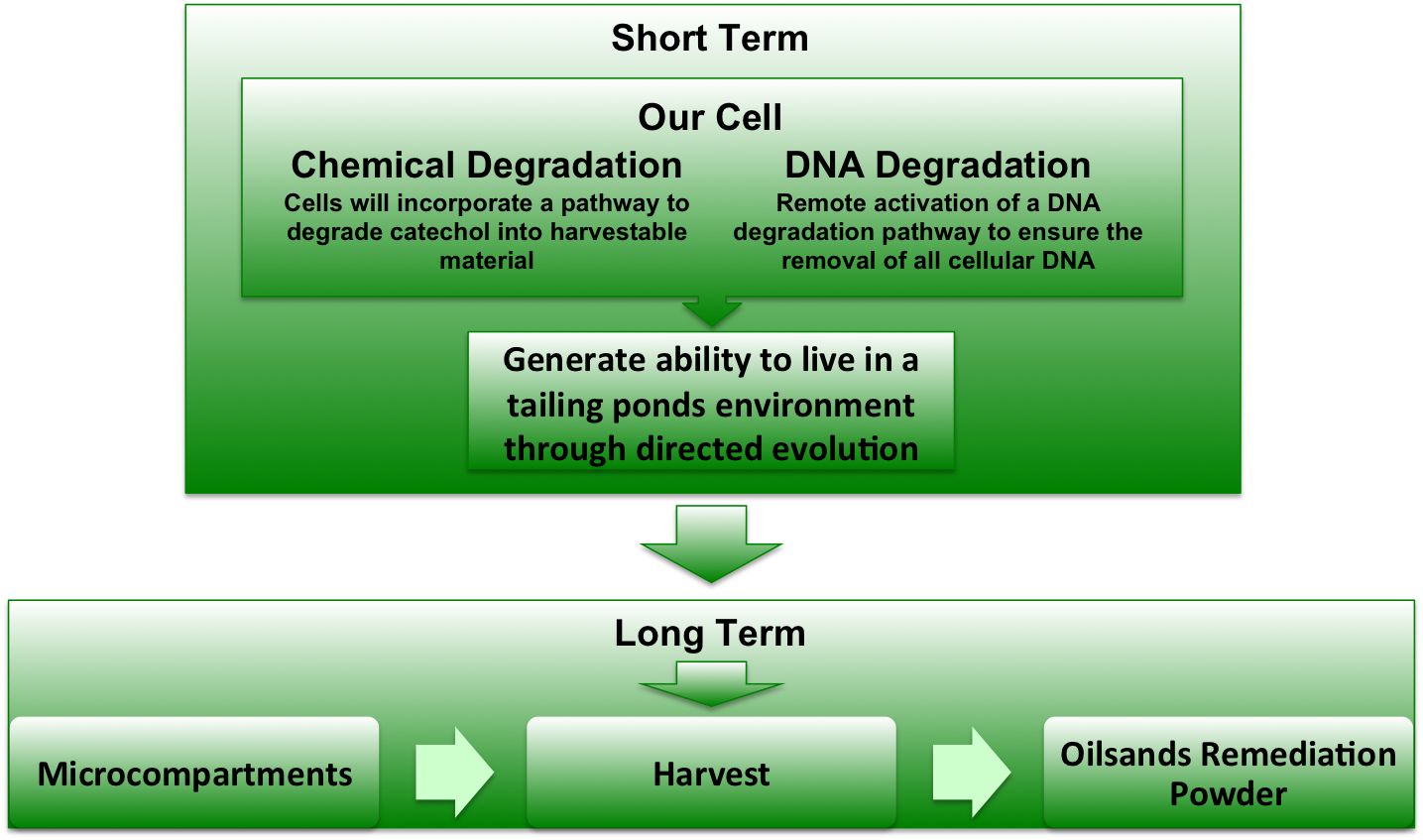
| + | [[Image:UofLProjectOverview.jpg|880px|Center]] | ||
| + | Figure 1. A depiction of the project overview. | ||
| + | ====<font color="white">Short-term Goals==== | ||
| - | === | + | We expect to have the <html><a href="https://2010.igem.org/Team:Lethbridge/Parts"><font color="#00DC00"> biobrick parts submitted</font></a></html> to the registry for all of the separate modules. Once we have the biobrick parts we will then be able to assemble the proposed system of the <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Catechol_Degradation"><font color="#00DC00"> catechol degradation pathway</font></a></html>, <html><a href="https://2010.igem.org/Team:Lethbridge/Project/DNA_Degradation"><font color="#00DC00"> induced cell death</font></a></html> and <html><a href="https://2010.igem.org/Team:Lethbridge/Project/Compartamentalization"><font color="#00DC00"> microcompartments</font></a></html>. Our short-term goal will be to provide characterized biobricks for the degradation of catechol in tailings. |
| + | ====<font color="white">Medium-term goals==== | ||
| + | We will accomplish the optimization of the system by selective evolution. To accomplish this we will use the tailings sample provided to optimize the assembled of the optimized catechol degradation pathway. | ||
| + | ====<font color="white">Long-term Goals==== | ||
| - | + | We will fully assemble the optimized modules for the tailings sample by selective evolution in preparation for use. The optimized pathway will be created into an easily administered powder which can be added to the tailing ponds which will degrade the catechol without addition of live organisms. This will allow for the tailings’ cleanup which will leave a smaller environmental footprint while simultaneously decreasing the time necessary for the tailings cleanup. | |
| + | <br> | ||
| + | <br> | ||
 "
"