Team:Kyoto/Project
From 2010.igem.org
Contents |
Abstract
Background
In synthetic biology field, biologists and bioengineers design and construct a variety of gene circuits to remedy a polluted environment. However, we can’t scatter genetically modified organisms on the environment. It is because that they may disturb the ecosystem. Therefore, to use them in the natural world, we need a system that genetically modified organisms die when they finish to remedy the polluted environment. In our project, we purpose to make the universal use system for cell death.
System of "Lysis Box"
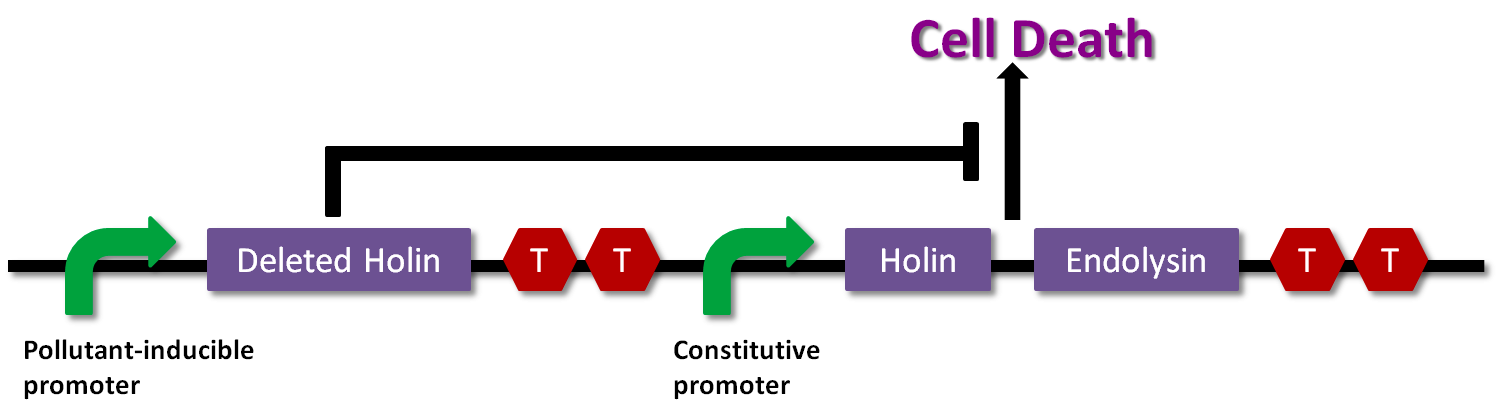
We use the lytic system of λphage. In this system, endolysin can access peptidoglycan of cell wall through the pores formed by holin and degrade it, leading the E.coli to die.
In “Lysis Box”, dominant-negative holin, from which a certain functional domain is deleted, inhibits the formation of the pores, inhibiting the cell death. The expression of dominant-negative holin depends on the induction of a pollutant. So as gene-modified organisms with this device degrades the pollutant little by little, the expression of dominant-negative holin becomes lower and lower. Finally, the inhibition of the cell death is lost and they die.
In this project, we aim to make “Lysis Box” and characterize it well from the viewpoint of the relationship between the expression level of dominant-negative holin and the cell death.
Introduction
Methods
Results
Discussion
Conclusion
Acknowledgments
Notes
References
- Berry, J., Summer, E.J., Struck, D.K., Young, R., "The final step in the phage infection cycle: the Rz and Rz1 lysis proteins link the inner and outer membranes." Mol Microbiol. vol. 70, issue. 2, Oct. 2008, pp. 341-51.
- Chang, C.Y., Nam, K., Young, R., “S gene expression and the timing of lysis by bacteriophage lambda.”, Journal of Bacteriology, vol. 177, Jun. 1995, pp. 3283-3294.
- Gründling, A., Manson, M.D., Young, R., "Holins kill without warning." Proceedings of the National Academy of Sciences of the United States of America, vol. 98, no. 16, Jul. 2001, pp. 9348-52.
- Kelly, J.R., Rubin, A.J., Davis, J.H., Ajo-Franklin, C.M., Cumbers, J., Czar, M.J., de Mora, K., Glieberman, A.L., Monie, D.D., Endy, D., "Measuring the activity of BioBrick promoters using an in vivo reference standard." Journal of biological engineering, Mar. 2009.
- Smith, D.L., Struck, D.K., Scholtz, J.M., Young, R., "Purification and biochemical characterization of the lambda holin.", Journal of Bacteriology, vol. 180, no. 9, May 1998, pp. 2531-40.
- White, R., Tran, T.A., Dankenbring, C.A., Deaton, J., Young, R., “The N-terminal transmembrane domain of lambda S is required for holin but not antiholin function.”, Journal of Bacteriology, vol. 192, Feb. 2010, pp. 725-733.
 "
"