Team:Imperial College London/Parts
From 2010.igem.org
| Line 1: | Line 1: | ||
| - | |||
{{:Team:Imperial_College_London/Templates/Header}} | {{:Team:Imperial_College_London/Templates/Header}} | ||
| Line 7: | Line 6: | ||
|'''We have contributed 23 parts to the Registry of Standard Biological Parts. We hope that other people will find the useful in the future. For more information on the individual parts, see below.''' | |'''We have contributed 23 parts to the Registry of Standard Biological Parts. We hope that other people will find the useful in the future. For more information on the individual parts, see below.''' | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 79: | Line 78: | ||
Sequence: ACGT etc. | Sequence: ACGT etc. | ||
| - | Short: pVeg Constitutive promoter for Veg locus from B. subtilis | + | Short: pVeg Constitutive promoter for Veg locus from ''B. subtilis'' |
Long: This part is identical to the sequence submitted by Imperial 2008 team, this part was produced from BBa_K143053 by PCR. PVeg is a constitutive promoter controlled by Sigma factor A. This promoter has two binding sites which leads to high expression of downstream genes. There is some evidence that the sporulation master regulator the spoOA can interact with pVeg although the mechanism is not known. | Long: This part is identical to the sequence submitted by Imperial 2008 team, this part was produced from BBa_K143053 by PCR. PVeg is a constitutive promoter controlled by Sigma factor A. This promoter has two binding sites which leads to high expression of downstream genes. There is some evidence that the sporulation master regulator the spoOA can interact with pVeg although the mechanism is not known. | ||
| Line 145: | Line 144: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 186: | Line 185: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 213: | Line 212: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 240: | Line 239: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 271: | Line 270: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 324: | Line 323: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 355: | Line 354: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 381: | Line 380: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 405: | Line 404: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 475: | Line 474: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 504: | Line 503: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 534: | Line 533: | ||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| Line 746: | Line 745: | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| - | | | + | |==**K316024 - K316023+dif - K143070-K143064**== |
| - | + | ||
| + | Name: 5'amye-diff-Pveg-spoVG-CAT-B0015 | ||
| + | |||
| + | Code: BBa_K316024 | ||
| + | |||
| + | Sequence: | ||
| + | |||
| + | Short: | ||
| + | |||
| + | Chloraphenicol resistance gene with dif and 5' amyE sequence | ||
| + | |||
| + | Long: | ||
| + | |||
| + | This part is identical to <bbpart>BBa_K143023</bbpart> except for the dif site<bbpart>BBa_K3160</bbpart> integrated behind the 5' amye <bbpart>BBa_K143001</bbpart> integration sequence. | ||
| + | |||
| + | Chloramphenicol acetyltransferase (CAT) <bbpart>BBa_J31005</bbpart> confers resistance to chloramphenicol, a common lab selection antibiotic. The 5' amyE <bbpart>BBa_K143001</bbpart> integration sequence allows integration into B. subtilis genome, which disrupts ability to breakdown starch. In order for integration to occur, 3' amyE integration sequence <bbpart>BBa_K143002</bbpart> is also required. | ||
| + | |||
| + | This part can be used as the 5' start of an amyE integration vector, the genes of interest can then be attached to the 3' end, followed by the 3' amye integratio sequwnce <bbpart>BBa_K143002</bbpart>. | ||
| + | |||
| + | Please see ‘Part Design’ section for design considerations and parts used. | ||
| + | |||
| + | Source: Biobrick parts <bbpart>BBa_K143070</bbpart> (which contains <bbpart>BBa_K143001</bbpart>, <bbpart>BBa_K143012</bbpart>, <bbpart>BBa_K143021</bbpart>) and <bbpart>BBa_K143064</bbpart> (which contains <bbpart>BBa_K143031</bbpart>, <bbpart>BBa_B0015</bbpart>) | ||
| + | |||
| + | Design: 'Reverse PCR' was used to amplify the whole vector, while adding the dif sites by primer extension. Pfu polymerase was used to ensure error-free replication. | ||
| + | |||
| + | References: Very helpful if you have them! | ||
| + | |||
| + | <biblio> | ||
| + | |||
| + | </biblio> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| - | | | + | |==K316025 - diff-K143002== |
| + | |||
| + | Name: oh noes!?!!?!!!! not this one!!! | ||
| + | |||
| + | Code: BBa_K316025 | ||
| + | |||
| + | Sequence: | ||
| + | |||
| + | Short: | ||
| + | |||
| + | Dif-PmeI site with 3' amyE integration sequence | ||
| + | |||
| + | Long: | ||
| + | |||
| + | This vector has been designed using the AmyE 5' and 3' integration sequences for integration into B.subtilis genome. This vector is identical to <bbpart>BBa_K316020</bbpart> aside from the LacI expression cassette made from <bbpart>BBa_K143053</bbpart> and <bbpart>BBa_K143062</bbpart>. | ||
| + | |||
| + | This construct was designed to test IPTG inducible <bbpart>BBa_K316009</bbpart>. | ||
| + | |||
| + | |||
| + | Source: Biobricks <bbpart>BBa_K143070</bbpart>, <bbpart>BBa_K316002</bbpart>, bbpart>BBa_K143064</bbpart> bbpart>BBa_K143053</bbpart> and <bbpart>BBa_K143062</bbpart> <bbpart>BBa_K143002</bbpart> and synthetic oligos <bbpart>BBa_K316014</bbpart> | ||
| + | |||
| + | Design: Standard biobrick assembly. everse PCR using Pfu to include the dif site after 5' amyE | ||
| + | |||
| + | References: Very helpful if you have them! | ||
| + | |||
| + | <biblio> | ||
| + | |||
| + | </biblio> | ||
| + | |||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| - | | | + | |==**K316027 -LacI testing vector K143001-k316002-K143053-K143065-K143053-K143062- K316002-K316014-K143002** == |
| + | |||
| + | Name:amyE complete vector 5'amyE-diff-Pveg-spoVG-CAT-B0015-diff-PmeI-3'amyE | ||
| + | |||
| + | Code: BBa_K316022 | ||
| + | |||
| + | Sequence: | ||
| + | |||
| + | Short: | ||
| + | |||
| + | B.subtilis transformation vector with LacI, targets Amylase locus | ||
| + | |||
| + | Long: | ||
| + | |||
| + | This vector has been designed using the amyE 5' and 3' integration sequences for integration into B.subtilis genome | ||
| + | |||
| + | '''AmyE locus''' | ||
| + | This vector has been designed using the amyE 5' <bbpart>BBa_K143008</bbpart> and 3 <bbpart>BBa_K143009</bbpart>' integration sequences for integration into ''B. subtilis'' genome. Insertion into the amyE locus provides a selection marker as the bacterium will no longer be able to breakdown starch. An iodine assay can be used to confirm integration. This phenotype makes the transformed bacterium considerably less likely to survive in natural environments. | ||
| + | |||
| + | '''Chloramphenicol Resistance''' | ||
| + | This vector also contains a positive selection marker, flanked by two dif sites. Chloramphenicol acetyltransferase <bbpart>BBa_J31005</bbpart> provides resistance to chloramphenicol antibiotic. Dif <bbpart>BBa_K316002</bbpart> sites allow excision of the antibiotic marker after integration, thus allowing the same marker to be used again or as a precaution against horizontal gene transfer. | ||
| + | |||
| + | '''Blunt end cloning site''' | ||
| + | PmeI restriction site <bbpart>BBa_K316013</bbpart> is designed as a cloning site. Due to the 8bp recognition sequence it is a rare site that can be used to cut the vector only once. | ||
| + | |||
| + | |||
| + | For more information about our project please visit our wiki [https://2010.igem.org/Team:Imperial_College_London] or take the tour [https://2010.igem.org/Team:Imperial_College_London/Tour/Page_One] to learn more about the project. | ||
| + | |||
| + | |||
| + | |||
| + | Source: Existing biobricks, <bbpart>BBa_K143070</bbpart>, <bbpart>BBa_K316002</bbpart>, <bbpart>BBa_K316014</bbpart> <bbpart>BBa_K143002</bbpart> | ||
| + | |||
| + | Design: This part is designed for integration into ''B. subtilis' genome at amyE locus, it contains Spectinomycin resistance selection marker and constitutively expresses LacI. PmeI can be used to ligate a gene of interest using blunt ended methods. | ||
| + | |||
| + | BBa_K143001 BBa_K316002 BBa_K143053 K143065 BBa_K143053 BBa_K1430562 BBa_K316014 BBa_K143002 | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | References: Very helpful if you have them! | ||
| + | |||
| + | <biblio> | ||
| + | |||
| + | </biblio> | ||
| + | |||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| - | | | + | |==**K316030 - LytC**== |
| + | |||
| + | Name: | ||
| + | |||
| + | Code: BBa_K316030 | ||
| + | |||
| + | Sequence: | ||
| + | |||
| + | Short: LytC cell wall anchoring protein for B. subtilis | ||
| + | |||
| + | Long: | ||
| + | The cell wall binding domin of lytC, a N-acetylmuramoyl-L-alanine amidase of B. subtilis, was isolated to be used as a cell wall anchor. As demonstrated by Kobayashi et al. 2000 this domin can be used to target catalytic domians of other proteins as well as peptides to the cell wall of B. subtilis whilst leaving their functionality intact. | ||
| + | |||
| + | Source: Genome PCR from B. subtilis genome (strain) | ||
| + | |||
| + | Design: Pfu DNA polymerase was used to minimise PCR errors | ||
| + | |||
| + | References: Very helpful if you have them! | ||
| + | |||
| + | <biblio> | ||
| + | |||
| + | </biblio> | ||
| + | |||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| - | | | + | |==**K316031 - K316001-LytC**== |
| - | + | ||
| - | + | Name: | |
| - | + | ||
| - | + | Code: BBa_K316031 | |
| - | + | ||
| + | Sequence: | ||
| + | |||
| + | Short: LytC cell wall anchoring protein with Pveg promoter | ||
| + | |||
| + | Long: | ||
| + | |||
| + | promoter - add links to both k316001 and K143012 | ||
| + | |||
| + | The cell wall binding domin of lytC, a N-acetylmuramoyl-L-alanine amidase of B. subtilis, was isolated to be used as a cell wall anchor. As demonstrated by Kobayashi et al. 2000 this domin can be used to target catalytic domians of other proteins as well as peptides to the cell wall of B. subtilis whilst leaving their functionality intact. | ||
| + | |||
| + | Source: Genome PCR from B. subtilis genome (strain) | ||
| + | |||
| + | Design: Pfu DNA polymerase was used to minimise PCR errors | ||
| + | |||
| + | References: Very helpful if you have them! | ||
| + | |||
| + | <biblio> | ||
| + | |||
| + | </biblio> | ||
| + | |||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| - | | | + | |==**K316031 - LytC with linker1**== |
| + | |||
| + | Name: LytC-linker1-TEV cleavable site-ComC | ||
| + | |||
| + | Code: BBa_K316031 | ||
| + | |||
| + | Sequence: | ||
| + | |||
| + | Short: B. subtilis Cell wall biniding domain with cleavable linker | ||
| + | |||
| + | Long: | ||
| + | We designed a protein that carries our signal peptide ComC out of the cell where a protease has access to it. The protease can then proceed to cleave ComC off the protein, allowing quorum sensing via the the ComCDE system to take place. One big problem we have to overcome is the cell wall that will obstruct the protease’s access to the protein carrying the signal peptide. The Detection module consists of a cell wall anchor, a signal peptide called ComC, and a linker that connects the two and is specifically designed to be cleaved by the protease we want to detect | ||
| + | |||
| + | Source: Genome PCR from B. subtilis genome (strain) with linker-TEV cleavable site-ComC synthesised and attached via a PmeI site within LytC gene | ||
| + | |||
| + | Design: Pfu DNA polymerase was used to minimise PCR errors | ||
| + | |||
| + | References: Very helpful if you have them! | ||
| + | |||
| + | <biblio> | ||
| + | |||
| + | </biblio> | ||
| + | |||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| - | | | + | |==K316032 - LytC== |
| - | + | ||
| - | + | Name: | |
| - | + | ||
| - | + | Code: BBa_K316032 | |
| - | + | ||
| + | Sequence: | ||
| + | |||
| + | Short: 3' strand encoded XylE under LacI activation | ||
| + | |||
| + | Long: | ||
| + | The cell wall binding domin of lytC, a N-acetylmuramoyl-L-alanine amidase of B. subtilis, was isolated to be used as a cell wall anchor. As demonstrated by Kobayashi et al. 2000 this domin can be used to target catalytic domians of other proteins as well as peptides to the cell wall of B. subtilis whilst leaving their functionality intact. | ||
| + | |||
| + | Source: Genome PCR from B. subtilis genome (strain) | ||
| + | |||
| + | Design: Pfu DNA polymerase was used to minimise PCR errors | ||
| + | |||
| + | References: Very helpful if you have them! | ||
| + | |||
| + | <biblio> | ||
| + | |||
| + | </biblio> | ||
| + | |||
| + | |||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| - | | | + | |==K316033 - LytC== |
| + | |||
| + | Name: LytC – Glycin Linker – TEV Cleavage Site – His-Tag – Stop Codon | ||
| + | |||
| + | Code: BBa_K316033 | ||
| + | |||
| + | Sequence: | ||
| + | |||
| + | Short: 3' strand encoded XylE under LacI activation | ||
| + | |||
| + | Long: | ||
| + | |||
| + | Introduction: This part was used to link the cell wall binding domain (CWB) of LytC, used in the detection module, with the quorum sensing peptide (AIP) as well as providing a cleavage site for a protease we want to detect. | ||
| + | LytC: The part carries part of LytC on its 5’ end. This was used to ligate the linker with LytC via an internal ACCI restriction site that occurs naturally (Link to LytC sequence). | ||
| + | Glycin Linker: The Linker separates the CWB and the AIP and creates space for the protease to access the cleavage site; it consists of two main sections. The first six amino acids (SRGSRA) were suggested to be used specifically with LytC (Yamamoto et al. 2003). The second section consists of a several glycin residues. | ||
| + | TEV Cleavage Site: This sequence forms the 3’ end of the linker and is directly attached to the 5’ end of the AIP. It is 18 amino acids (GGGGENLYFQGGKLGGGG) long and was designed to be efficiently cleaved by the TEV protease, as well as being codon-optimised for expression in ''B. subtilis''. | ||
| + | His-Tag: To be able to purify the protein for testing, we attached a His-Tag on our linker-AIP peptide. As it would probably interfere with recognition of the AIP by the receptor it has to be removed from the final construct. | ||
| + | Stop Codon: In order to end translation a double stop codon was put in place. | ||
| + | |||
| + | |||
| + | Source: Genome PCR from B. subtilis genome (strain) | ||
| + | |||
| + | Design: Pfu DNA polymerase was used to minimise PCR errors | ||
| + | |||
| + | References: Very helpful if you have them! | ||
| + | |||
| + | <biblio> | ||
| + | |||
| + | </biblio> | ||
| + | |||
| + | ==K316040 - sRBS== | ||
| + | |||
| + | Name: | ||
| + | |||
| + | Code: BBa_K316040 | ||
| + | |||
| + | Sequence: | ||
| + | |||
| + | Short: Synthetic RBS for LytC cell wall anchoring protein | ||
| + | |||
| + | Long: Optimal Ribosome Binding Site (RBS) designed for high expression levels in B. subtilis. Opimised using Vogit Lab RBS calculator v 1.0 [http://voigtlab.ucsf.edu/software/] this software is based on | ||
| + | |||
| + | |||
| + | Source: Genome PCR from B. subtilis genome (strain) | ||
| + | |||
| + | Design: Pfu DNA polymerase was used to minimise PCR errors | ||
| + | |||
| + | References: Very helpful if you have them! | ||
| + | |||
|} | |} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Parts | ||
|- | |- | ||
| | | | ||
|} | |} | ||
Revision as of 00:54, 27 October 2010
| Parts |
| We have contributed 23 parts to the Registry of Standard Biological Parts. We hope that other people will find the useful in the future. For more information on the individual parts, see below. |
| Parts |
| ==**K316012 - 3' K316000-sRBS-TEV**==
Name: 3' coding, Enhanced LacI inducible promoter, synthetic RBS, TEV Code: BBa_K316000 Sequence: Short: 3' coding TEV protease under control of enhanced Lac operon Long: TEV protease S219P autocatalysis resistant variant Introduction : This is the nuclear inclusion protease, endogenous to Tobacco Etch Virus and is used in the late lifecycle to cleave polyprotein precursors. The recognition sequence is ENLYFQG/S [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6W9V-45PMGK3-9P&_user=217827&_coverDate=02%2F01%2F1994&_rdoc=1&_fmt=high&_orig=search&_origin=search&_sort=d&_docanchor=&view=c&_acct=C000011279&_version=1&_urlVersion=0&_userid=217827&md5=e075aad3a349720ad9484095d01a65be&searchtype=a]] between QG or QSDue to it’s stringent sequence specificity, TEV is commonly used to cleave genetically engineered proteins. Uses: TEV proteinase is used to cleave fusion proteins. It is useful due to its high degree of specificity [[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6W9V-45PMGK3-9P&_user=217827&_coverDate=02%2F01%2F1994&_rdoc=1&_fmt=high&_orig=search&_origin=search&_sort=d&_docanchor=&view=c&_acct=C000011279&_version=1&_urlVersion=0&_userid=217827&md5=e075aad3a349720ad9484095d01a65be&searchtype=a]] and potential to be used in vivo or in vitro applications.
Wild type TEV protease also cleaves itself at Met 218 and Ser 219 [[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WXR-45R86R6-5X&_user=7635175&_coverDate=06%2F20%2F1995&_rdoc=1&_fmt=high&_orig=search&_origin=search&_sort=d&_docanchor=&view=c&_acct=C000011279&_version=1&_urlVersion=0&_userid=7635175&md5=0a1521e783c0bd4126b145f3f6d766d4&searchtype=a]] . This leads to auto-inactivation of the TEV protease and progressive loss of activity of the protein. The rate of inactivation is proportional to the concentration of protease http://peds.oxfordjournals.org/content/14/12/993.long More stable Mutants have been produced by single amino acid substitutions S219V (AGC(serine) to GTG(valine) and S219P (AGC(serine) to CCG(proline) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency Table I. Kinetic parameters for wild-type and mutant TEV proteases with the peptide substrate TENLYFQSGTRR-NH2. Enzyme Km (mM) kcat (s−1) kcat/Km (mM−1 s−1) Wild-type 0.061 ± 0.010 0.16 ± 0.01 2.62 ± 0.46 S219V* 0.041 ± 0.010 0.19 ± 0.01 4.63 ± 1.16 S219P* 0.066 ± 0.008 0.09 ± 0.01 1.36 ± 0.22 S219P* - virtually imperivious to autocatalysis S219V* - retains same activity as wild type Full article can be seen here http://peds.oxfordjournals.org/content/14/12/993.long
Design: The part was produced by nucleotide synthesis by mwg – eurofins References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316001 - Pveg**==
Name: Pveg promoter Code: BBa_K316001 Sequence: ACGT etc. Short: pVeg Constitutive promoter for Veg locus from B. subtilis Long: This part is identical to the sequence submitted by Imperial 2008 team, this part was produced from BBa_K143053 by PCR. PVeg is a constitutive promoter controlled by Sigma factor A. This promoter has two binding sites which leads to high expression of downstream genes. There is some evidence that the sporulation master regulator the spoOA can interact with pVeg although the mechanism is not known. Source: PCR from existing biobrick K143053 using Pfu polymerase II Design: PCR using Pfu polymerase to avoid mutations References: Very helpful if you have them! <biblio> </biblio> |
| Parts | |
| ==**K316002 - dif**==
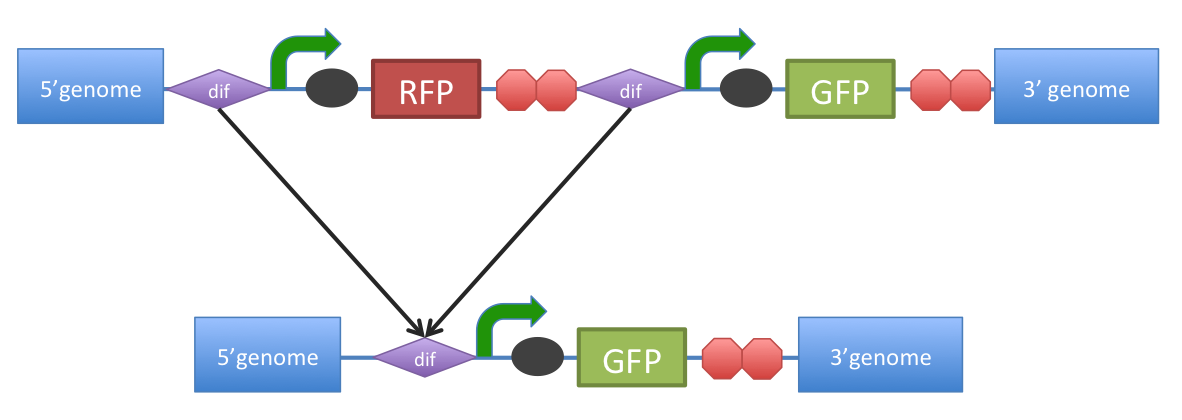
Name: B. subtilis dif excision site Code: BBa_K316002 Sequence: Short: B. subtilis dif: sequence-specific recombinase target site Long:
B. subtilis dif: sequence-specific recombinase recognition siteCells with circular chromosomes, recombinatorial repair and homologous recombination can generate multimeric chromosomes 1. ‘’Dif’’ sites are part of a system to ensure that multimeric chromosomes can be separated to monomers, which is required for proper sharing of genetic material between daughter cells. In ‘’B. subtilis’’ the tyrosine family recombinases such as RipX and CodV mediate the separation at 28-bp sequence Bs’’dif’’ 1. The site-specific recombinases are able to recognize two directly repeated ’’dif’’ sites and excise the fragment flanked by the two sites 2.
http://partsregistry.org/Image:DifExcision.PNG Figure 1. Removal of a specific gene from a genome integrated construct.The site-specific recombinases, endogenous to ‘’B. subtilis’’ strains are able to recognise Bs’’dif’’ sites and recombine out the strand of DNA directly flanked by the two sites. Recombination leaves a single dif site. The construct was previously engineered to homologously recobine into the genome of ‘’B. subtilis’’. Integration sequences such as amyE <bbpart>BBa_J143001</bbpart>, <bbpart>BBa_J143002</bbpart> can be used to achieve this. Source: The dif sites were made by annealing synthestised oligoes. Design: The dif site was made by oligos designed to make overhangs for EcoRI and SpeI ( and ) or XbaI and PstI ( and ) to be used in standard Biobrick or 3A cloning. References: Very helpful if you have them! References<biblio>
</biblio>References |
| Parts |
| ==**K316003 - XylE-B0014** ==
Name: XylE, Double Terminator Code: BBa_S04510 Sequence: Short: XylE - catechol 2,3-dioxygenase from P.putida with terminator Long: Catechol or catechol 2,3-dioxygenase + O(2) is converted by a ring cleavage into 2-hydroxymuconate semialdehyde which is the toxic and bright yellow-coloured substrate. This is a key enzyme in many (soil) bacterial species used for the degradation of aromatic compounds. The catechol 2,3-dioxygenase (pdb id: 1MPY[http://www.ebi.ac.uk/pdbsum/1mpy]) used here itself originating from Pseudomonas putida is a homotetramer of C230 monomers. The tetramerization interactions position a ferrous ion critical for enzymatic activity. It has been deduced that intersubunit interaction is essential to produce a functioning enzyme after performing N and C terminal modifications on the monomer. Coming together the subunits generate an active site. The reaction itself takes place within seconds after the addition by Pasteur pipette or spraying of catechol at a 100mM stock solution diluted with DDH20 (used by our lab.) The toxic byproduct is thought to interfere with cell wall integrity and cellular machinery such that exposed cells gradually die. Catechol 2,3-dioxygenase Source: XylE was obtained from the registry <bbpart>BBa_K118021</bbpart>, the terminator is <bbpart>BBa_B0014</bbpart>. XylE is naturally found in Pseudomonas putida. Design: The parts were put together using standard assembly [http://partsregistry.org/Assembly:Standard_assembly]
|
| Parts |
| ==**K316004 - J23101-XylE-B0014** ==
Name: Constitutive E-coli promoter, XylE, double terminator Code: BBa_K316004 Sequence: Short: Functional XylE under J23101 promoter, with B0014 terminator Long: <bbpart>BBa_J23101</bbpart> combined with <bbpart>BBa_S04510</bbpart>for easy characterisation of gene activity (catechol breakdown into 2,4 hydroxyaminobutane yellow product) under a standard E.coli promoter Source: Biobrick parts <bbpart>BBa_J23101</bbpart> and <bbpart>BBa_S04510</bbpart> Design: Parts were assembled in two steps using standard biobrick assembly. References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316005 - K316001-XylE-B0014**==
Name: Pveg-XylE-Double Terminator Code: BBa_K316005 Sequence: Short: Functional XylE under Pveg promoter, with B0014 terminator Long: Enables characterization and comparison with alternative promoters e.g. <bbpart>BBa_J23101</bbpart> Designed for characterisation of XylE in Bacillus subtilis where Pveg is becoming a standard promoter Source: Biobrick parts <bbpart>BBa_J33204</bbpart> <bbpart>BBa_B0014</bbpart> Design: Parts were assembled in two steps using standard biobrick assembly. References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316006 - 5'hisGFP-XylE**==
Name: 5'his tagged-GFP-XylE fusion protein Code: BBa_K316006 Sequence: Short: 5'His-tagged GFP linked to XylE by TEV cleavable linker Long: Constructed to be combined with promoter and terminator. The GFP <bbpart>BBa_E0040</bbpart> is linked to XylE <bbpart>BBa_J33204</bbpart> monomer subunit by a GGGSGGGS linker with the aim to render the enzyme inactive, via preventing tetramerization (it’s functional form). Please see ‘Part Design’ section for design considerations and parts used. Source: Existing biobrick parts with modifications using PCR primer extension Design: 'PCR primer extension - exact methods our wiki [1] References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316007 - K143053-GFP-XylE**==
Name: B. subtilis promoter Pveg and strong RBS spoVG <bbpart>BBa_K143053</bbpart>, 5'His tagged GFP-linker-XylE fusion protein Code: BBa_K316007 Sequence: Short: Cleavable GFP-XylE protein under Pveg promoter Long: This construct is designed so that the XylE activity is substantially reduced untill such a time when a TEV protease is added to the system and transcribed. TEV protease cleavable linker is positioned between the two proteins. Once the linker is cleaved, XylE is free to tetramerise and assume full activity. GFP is His tagged at the 5' end to facilitate purificaiton for in-vitro assays. Source: Pveg and spoVG biobricks <bbpart>BBa_K143053</bbpart> added to GFP-XylE construct <bbpart>BBa_K316004</bbpart>. Design: Standard biobrick cloning of intermediary parts add part numbers <bbpart>BBa_K143053</bbpart> References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316008 - K143053-5'hisGFP-linker-XylE-B0014**==
Name: B. subtilis promoter Pveg and strong RBS spoVG <bbpart>BBa_K143053</bbpart>, 5'His tagged GFP-linker-XylE fusion protein <bbpart>BBa_K316005</bbpart>, double terminator <bbpart>BBa_B0014</bbpart> Code: BBa_K316008 Sequence: Short: Cleavable GFP-XylE fusion with Pveg promoter and terminator Long: This construct is designed so that the XylE activity is substantially reduced untill such a time when a TEV protease is added to the system and transcribed. TEV protease cleavable linker is positioned between the two proteins. Once the linker is cleaved, XylE is free to tetramerise and assume full activity. GFP is His tagged at the 5' end to facilitate purificaiton for in-vitro assays. Terminator <bbpart>BBa_B0014</bbpart> has been added to comply with Biobrick standards. This particular terminator is stronger and is different from <bbpart>BBa_B0015</bbpart>. Source: Existing Biobricks <bbpart>BBa_K143053</bbpart>, <bbpart>BBa_K316005</bbpart>, <bbpart>BBa_B0015</bbpart> Design: Standard Biobrick assembly References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316009 -TEV-sRBS-K316000 -K143053-5'hisGFP-linker-XylE-B0014**==
Name: minus strand encoded - LacI inducible TEV and plus strand 5'his tagged-GFP-linker-XylE construct under constitutive Pveg promoter and spoVG RBS <bbpart>BBa_K143053</bbpart> Code: BBa_K316009 Sequence: Short: LacI inducible Fast Response Module, using cleavable XylE Long: 5' his tagged GFP-TEV linker-XylE construct is pre-made in the cell under constitutive promoter. Both Pveg promoter and spoVG RBS are best suited for maximal expression in B.subtilis. While GFP is attached to the XylE monomer via the TEV cleavable linker, the catalytic activity is low. Transcription of TEV protease allows cleavage of the linker between GFP and XylE, thus XylE is free to tetramerise into a fully functional enzyme. XylE is then able to act as described in <bbpart>BBa_k316004</bbpart> Please see 'Part Design' section for design considerations and parts used. Source: Made from parts - <bbpart>BBa_k316012</bbpart> and <bbpart>BBa_k316008</bbpart> Design: Standard biobrick assembly References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==K316010 -** 3' encoded K316000 - XylE**==
Name: LacI inducible XylE Code: BBa_K316010 Sequence: Short: 3' strand coding XylE under LacI activation Long: This part contains XylE under synthetic hyperspank promoter <bbpart>BBa_K143000</bbpart> LacI and IPTG are required for repression and activation of expression. As transformation of B subtilis often requires genomic integration, read-through from upstream genomic regions can become an issue. For the purposes of characterisation, the part was made on the minus strand to reduce background transcription. Read-through may provide an additional benefit as low quantities of anti-sence RNA may reduce basal expression sometimes seen in LacI systems. (although there is some evidence background is usually due to readthrough if have ref) Source: Design: the part contains a synthetic RBS optimised for high expression in B subtilis, calculated using RBS calculator [ link ] References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==K316011 - Flipped Xyle==
Name: Flipped XylE Code: BBa_K316011 Sequence:
Long: This part contains XylE coding on the 3' strand. In many biological settings there may be read-through caused by upstream elements, however this can often be unidirectional. In settings where readthrough from 5' direction is expected to be much stronger than 3' direction, it may is advantageous to use 3' coding sequence instead of traditional 5'. For the purposes of characterisation, the part was made on the minus strand to reduce background transcription. Read-through from 5' direction may provide an additional benefit. Low quantities of anti-sence RNA may be produced as the result of 5' readthrough, which may in turn reduce basal expression sometimes seen in LacI systems. Source: This part is a modified existing biobrick <bbpart>BBa_J33204</bbpart> Design: Part was designed using several rounds of PCR, using Pfu polymerase to avoid mutations. For more information, please check our wiki[2] |
| Parts |
| ==**K316000 - Pehs** ==
Name: Enhanced hyperspank promoter Code: BBa_K316000 Sequence: Short: 3' coding Enhanced LacI-hyperspank promoter Long: This part is a modified version of hyper-spank promoter for B.subtilis <bbpart>BBa_K143015</bbpart>. Hyper-spank promoter is repressed by transcriptional repressor LacI <bbpart>BBa_K143033</bbpart> and can be induced by addition of Isopropyl β-D-1-thiogalactopyranoside (IPTG). Constitutive expression of LacI is required for repression. Promoter Design The position and sequence of LacI binding was designed using existing knowledge. The stochastic nature of transcriptional repressors usually leads to background transcription. In order to minimise background the binding sites and the distance between them have been optimised. Stronger binding The natural LacI operator has 3 binding sites, all of which have variations in the binding sequences. Perfectly symmetric binding sequence was shown to have10-fold higher binding compared to wild type sequences. The aattgtgagc gctcacaatt sequence has been shown to be optimal for LacI binding Muller 1996 Oehler 1994. Optimal distance Due to the tetrameric nature of LacI it can simulataneously bind to multiple regions in the genome. Binding at multple sites can produce much stronger repression (muller 1996) by increasing local LacI concentrations. Due to the helical nature of DNA the distance between the operator sites plays an important role in the strength of repression. Maximal repression at 70.5bp, second strongest at 92.5bp and third at 115.5bp
Design: Designed for minimal basal transcription by altering binding site sequence and distance. References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316013 - PmeI cutting site** ==
Name: PmeI recognition sequence Code: BBa_K316014 Sequence: Short: 8bp recognition sequence for PmeI restriction endonuclease Long: Information about PmeI restriction endonuclease is available at [http://www.neb.com/nebecomm/products/productR0560.asp]. The recognition site is a 8bp sequence GTTTAAAC. Pme produces a blunt cut after GTTT. Source: This is a planning part. The sequence was combined with <bbpart>BBa_K143000</bbpart> to produce <bbpart>BBa_K143014</bbpart> in construction of B. subtilis genome integration vectors Design: This is a planning part. The sequence was produced from single stranded primer oligos. References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316014 - K316000-K316013** ==
Name: Dif Pme Code: BBa_K316014 Sequence: Short: Dif sequence followed by PmeI recognition site Long: This composite part of <bbpart>BBa_K143000</bbpart> and <bbpart>BBa_K143013</bbpart>. The dif site can be used in conjunction with another dif site in another part of the vector to remove a sequence between the two dif sites. PmeI site can be used for blunt end cloning of a DNA sequence behind the dif site. Source: oligonucleotide synthesis Design: This part was designed to be cloned using standard biobrick methods. Two single stranded, synthetic oligos were annealed to produce double stranded DNA sequence with single stranded overhangs identical to the product of digestion by XbaI and SpeI. Thus compatible with biobrick cloning methods. References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316015 - ComD**==
Name: ComD Receptor Code: BBa_K316015 Sequence: Short: ComD receptor for CSP-1 (S. pneumoniae). Phosphorylates ComE Long: ComD is the histidine kinase of a two-component signal transduction signaling pathway in a Streptococcus pneumoniae quorum sensing system. It detects the linear autoinducing peptide (AIP) called Competence-Stimulating Peptide-1 (CSP-1) which is coded for by the ComC gene. Upon detection of CSP-1, ComD will autophosphoylate and then phosphorylate and activate the response regulator, ComE Source: Streptococcus pneumoniae, AAC44896.1, codon optimised for expression in B.subtilis Design: the part contains a synthetic RBS optimised for high expression in B subtilis, calculated using RBS calculator [ link ] References: Info on threshold levels of CSP-1 etc http://ukpmc.ac.uk/backend/ptpmcrender.cgi?accid=PMC40587&blobtype=pdf <biblio> </biblio> |
| Parts |
| ==**K316016 -ComE**==
Name: LacI inducible XylE Code: BBa_K316016 Sequence: Short: Activates transcription of target genes when phosphorylated. Long: ComE is the response regulator of a two-component signal transduction signaling pathway in a Streptococcus pneumoniae quorum sensing system. It is phosphorylate and activated by ligand-bound ComD. It will then induce transcription of any gene which has a ComE binding site in its promoter.
Design: Synthetic RBS to optimise translation in B. subtilis as part of poly-mRNA containing ComD upstream References: Designing a ComE binding site: http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00584.x/full Dimerisation of ComE and more info on binding site: http://jb.asm.org/cgi/content/full/186/10/3078
</biblio> |
| Parts |
| ==**K316017 -TEV+B316000**==
Name: LacI inducible TEV protease
LacI operator controlled TEV protease Long
Please note all the sequences of this part are reversed to code on 3' strand
The part was produced by nucleotide synthesis by mwg – eurofins. The synthetic RBS was designed to work with <bbpart>BBa_K316000</bbpart>.
|
| Parts |
| ==**K316018 -Com CDE forward==
Name: ComE responsive promoter
ComE responsive promoter Long ComE binds to a specific sequence leading to activation of trancription.
The part was produced by nucleotide synthesis by mwg – eurofins. The synthetic RBS was designed to work with <bbpart>BBa_K316000</bbpart>.
|
| Parts |
| ==**K316020 -pyrD complete vector-K143008-K316002-K143053-K143065-K316014-K143009**==
Name: B. subtilis genome integration vector into pyrD locus Code: BBa_K316020 Sequence: Short: B. subtilis genome integration vector into pyrD locus Long: PyrD This vector has been designed using the Pyrd 5' <bbpart>BBa_K143001</bbpart> and 3' <bbpart>BBa_K143002</bbpart> integration sequences for integration into B. subtilis genome. Insertion into the pyrD locus provides a negative selection marker as the bacterium will no longer be able to synthesise uracil. Thus medium supplement of 40ug/ml is required for growth. This phenotype makes the transformed bacterium considerably less likely to survive in natural environments. Spectinomycin Resistance This vector also contains a positive selection marker, flanked by two dif sites. Aad9 <bbpart>BBa_K143065</bbpart> provides resistance to spectinomycin antibiotic. Dif <bbpart>BBa_K316002</bbpart> sites allow excision of the antibiotic marker after integration, thus allowing the same marker to be used again or as a precaution against horizontal gene transfer. Blunt end cloning site PmeI restriction site <bbpart>BBa_K316013</bbpart> is designed as a cloning site. Due to the 8bp recognition sequence it is a rare site that can be used to cut the vector only once.
Source: Existing biobricks, <bbpart>BBa_K143008</bbpart>, <bbpart>BBa_K316002</bbpart>, <bbpart>BBa_K143053</bbpart> <bbpart>BBa_K143065</bbpart> <bbpart>BBa_K316014</bbpart> <bbpart>BBa_K143009</bbpart> Design: Using standard assembly of biobricks BBa_K143008 BBa_K316002 BBa_K143053 BBa_K143065 BBa_K316014 BBa_K143009 References: Very helpful if you have them! <biblio> </biblio> **BBa_K316021 BBa_K143053 BBa_K143065**Name: Pveg-spoVG-Spec-B0015 Code: BBa_K316021 Sequence: Short: Spectinomycin resistance with promoter for B.subtilis Long: For reliable expression in B. subtilis the strongest constitutive promoter Pveg and strong ribosome binding site spoVG <bbpart>BBa_K143053</bbpart> were combined with spectinomycin resistance gene with terminator <bbpart>BBa_J143065</bbpart>. Please see ‘Part Design’ section for design considerations and parts used.
Source: Existing biobricks <bbpart>BBa_K143053</bbpart>, <bbpart>BBa_J143065</bbpart> Design considerations: Standard biobrick assembly [http://partsregistry.org/Assembly:Standard_assembly] References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316022 - K143001-k316002-K143053-K143065-K316002-K316014-K143002** ==
Name:amyE complete vector 5'amyE-diff-Pveg-spoVG-CAT-B0015-diff-PmeI-3'amyE Code: BBa_K316022 Sequence: Short: B.subtilis transformation vector, targets Amylase locus Long: This vector has been designed using the amyE 5' and 3' integration sequences for integration into B.subtilis genome AmyE locus This vector has been designed using the amyE 5' <bbpart>BBa_K143008</bbpart> and 3 <bbpart>BBa_K143009</bbpart>' integration sequences for integration into B. subtilis genome. Insertion into the amyE locus provides a selection marker as the bacterium will no longer be able to breakdown starch. An iodine assay can be used to confirm integration. This phenotype makes the transformed bacterium considerably less likely to survive in natural environments. Chloramphenicol Resistance This vector also contains a positive selection marker, flanked by two dif sites. Chloramphenicol acetyltransferase <bbpart>BBa_J31005</bbpart> provides resistance to chloramphenicol antibiotic. Dif <bbpart>BBa_K316002</bbpart> sites allow excision of the antibiotic marker after integration, thus allowing the same marker to be used again or as a precaution against horizontal gene transfer. Blunt end cloning site PmeI restriction site <bbpart>BBa_K316013</bbpart> is designed as a cloning site. Due to the 8bp recognition sequence it is a rare site that can be used to cut the vector only once.
Source: Existing biobricks, <bbpart>BBa_K143070</bbpart>, <bbpart>BBa_K316002</bbpart>, <bbpart>BBa_K316014</bbpart> <bbpart>BBa_K143002</bbpart> Design: the part contains a synthetic RBS optimised for high expression in B subtilis, calculated using RBS calculator [ link ] BBa_K143001 BBa_K316002 BBa_K143052 BBa_J31005 BBa_K316014 BBa_K143002 BBa_K143001 BBa_K143052 BBa_J31005 BBa_K143002 References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316024 - K316023+dif - K143070-K143064**==
Name: 5'amye-diff-Pveg-spoVG-CAT-B0015 Code: BBa_K316024 Sequence: Short: Chloraphenicol resistance gene with dif and 5' amyE sequence Long: This part is identical to <bbpart>BBa_K143023</bbpart> except for the dif site<bbpart>BBa_K3160</bbpart> integrated behind the 5' amye <bbpart>BBa_K143001</bbpart> integration sequence. Chloramphenicol acetyltransferase (CAT) <bbpart>BBa_J31005</bbpart> confers resistance to chloramphenicol, a common lab selection antibiotic. The 5' amyE <bbpart>BBa_K143001</bbpart> integration sequence allows integration into B. subtilis genome, which disrupts ability to breakdown starch. In order for integration to occur, 3' amyE integration sequence <bbpart>BBa_K143002</bbpart> is also required. This part can be used as the 5' start of an amyE integration vector, the genes of interest can then be attached to the 3' end, followed by the 3' amye integratio sequwnce <bbpart>BBa_K143002</bbpart>. Please see ‘Part Design’ section for design considerations and parts used. Source: Biobrick parts <bbpart>BBa_K143070</bbpart> (which contains <bbpart>BBa_K143001</bbpart>, <bbpart>BBa_K143012</bbpart>, <bbpart>BBa_K143021</bbpart>) and <bbpart>BBa_K143064</bbpart> (which contains <bbpart>BBa_K143031</bbpart>, <bbpart>BBa_B0015</bbpart>) Design: 'Reverse PCR' was used to amplify the whole vector, while adding the dif sites by primer extension. Pfu polymerase was used to ensure error-free replication. References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==K316025 - diff-K143002==
Name: oh noes!?!!?!!!! not this one!!! Code: BBa_K316025 Sequence: Short: Dif-PmeI site with 3' amyE integration sequence Long: This vector has been designed using the AmyE 5' and 3' integration sequences for integration into B.subtilis genome. This vector is identical to <bbpart>BBa_K316020</bbpart> aside from the LacI expression cassette made from <bbpart>BBa_K143053</bbpart> and <bbpart>BBa_K143062</bbpart>. This construct was designed to test IPTG inducible <bbpart>BBa_K316009</bbpart>.
Design: Standard biobrick assembly. everse PCR using Pfu to include the dif site after 5' amyE References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316027 -LacI testing vector K143001-k316002-K143053-K143065-K143053-K143062- K316002-K316014-K143002** ==
Name:amyE complete vector 5'amyE-diff-Pveg-spoVG-CAT-B0015-diff-PmeI-3'amyE Code: BBa_K316022 Sequence: Short: B.subtilis transformation vector with LacI, targets Amylase locus Long: This vector has been designed using the amyE 5' and 3' integration sequences for integration into B.subtilis genome AmyE locus This vector has been designed using the amyE 5' <bbpart>BBa_K143008</bbpart> and 3 <bbpart>BBa_K143009</bbpart>' integration sequences for integration into B. subtilis genome. Insertion into the amyE locus provides a selection marker as the bacterium will no longer be able to breakdown starch. An iodine assay can be used to confirm integration. This phenotype makes the transformed bacterium considerably less likely to survive in natural environments. Chloramphenicol Resistance This vector also contains a positive selection marker, flanked by two dif sites. Chloramphenicol acetyltransferase <bbpart>BBa_J31005</bbpart> provides resistance to chloramphenicol antibiotic. Dif <bbpart>BBa_K316002</bbpart> sites allow excision of the antibiotic marker after integration, thus allowing the same marker to be used again or as a precaution against horizontal gene transfer. Blunt end cloning site PmeI restriction site <bbpart>BBa_K316013</bbpart> is designed as a cloning site. Due to the 8bp recognition sequence it is a rare site that can be used to cut the vector only once.
Source: Existing biobricks, <bbpart>BBa_K143070</bbpart>, <bbpart>BBa_K316002</bbpart>, <bbpart>BBa_K316014</bbpart> <bbpart>BBa_K143002</bbpart> Design: This part is designed for integration into B. subtilis' genome at amyE locus, it contains Spectinomycin resistance selection marker and constitutively expresses LacI. PmeI can be used to ligate a gene of interest using blunt ended methods. BBa_K143001 BBa_K316002 BBa_K143053 K143065 BBa_K143053 BBa_K1430562 BBa_K316014 BBa_K143002
<biblio> </biblio> |
| Parts |
| ==**K316030 - LytC**==
Name: Code: BBa_K316030 Sequence: Short: LytC cell wall anchoring protein for B. subtilis Long: The cell wall binding domin of lytC, a N-acetylmuramoyl-L-alanine amidase of B. subtilis, was isolated to be used as a cell wall anchor. As demonstrated by Kobayashi et al. 2000 this domin can be used to target catalytic domians of other proteins as well as peptides to the cell wall of B. subtilis whilst leaving their functionality intact. Source: Genome PCR from B. subtilis genome (strain) Design: Pfu DNA polymerase was used to minimise PCR errors References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316031 - K316001-LytC**==
Name: Code: BBa_K316031 Sequence: Short: LytC cell wall anchoring protein with Pveg promoter Long: promoter - add links to both k316001 and K143012 The cell wall binding domin of lytC, a N-acetylmuramoyl-L-alanine amidase of B. subtilis, was isolated to be used as a cell wall anchor. As demonstrated by Kobayashi et al. 2000 this domin can be used to target catalytic domians of other proteins as well as peptides to the cell wall of B. subtilis whilst leaving their functionality intact. Source: Genome PCR from B. subtilis genome (strain) Design: Pfu DNA polymerase was used to minimise PCR errors References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==**K316031 - LytC with linker1**==
Name: LytC-linker1-TEV cleavable site-ComC Code: BBa_K316031 Sequence: Short: B. subtilis Cell wall biniding domain with cleavable linker Long: We designed a protein that carries our signal peptide ComC out of the cell where a protease has access to it. The protease can then proceed to cleave ComC off the protein, allowing quorum sensing via the the ComCDE system to take place. One big problem we have to overcome is the cell wall that will obstruct the protease’s access to the protein carrying the signal peptide. The Detection module consists of a cell wall anchor, a signal peptide called ComC, and a linker that connects the two and is specifically designed to be cleaved by the protease we want to detect Source: Genome PCR from B. subtilis genome (strain) with linker-TEV cleavable site-ComC synthesised and attached via a PmeI site within LytC gene Design: Pfu DNA polymerase was used to minimise PCR errors References: Very helpful if you have them! <biblio> </biblio> |
| Parts |
| ==K316032 - LytC==
Name: Code: BBa_K316032 Sequence: Short: 3' strand encoded XylE under LacI activation Long: The cell wall binding domin of lytC, a N-acetylmuramoyl-L-alanine amidase of B. subtilis, was isolated to be used as a cell wall anchor. As demonstrated by Kobayashi et al. 2000 this domin can be used to target catalytic domians of other proteins as well as peptides to the cell wall of B. subtilis whilst leaving their functionality intact. Source: Genome PCR from B. subtilis genome (strain) Design: Pfu DNA polymerase was used to minimise PCR errors References: Very helpful if you have them! <biblio> </biblio>
|
| Parts |
| ==K316033 - LytC==
Name: LytC – Glycin Linker – TEV Cleavage Site – His-Tag – Stop Codon Code: BBa_K316033 Sequence: Short: 3' strand encoded XylE under LacI activation Long: Introduction: This part was used to link the cell wall binding domain (CWB) of LytC, used in the detection module, with the quorum sensing peptide (AIP) as well as providing a cleavage site for a protease we want to detect. LytC: The part carries part of LytC on its 5’ end. This was used to ligate the linker with LytC via an internal ACCI restriction site that occurs naturally (Link to LytC sequence). Glycin Linker: The Linker separates the CWB and the AIP and creates space for the protease to access the cleavage site; it consists of two main sections. The first six amino acids (SRGSRA) were suggested to be used specifically with LytC (Yamamoto et al. 2003). The second section consists of a several glycin residues. TEV Cleavage Site: This sequence forms the 3’ end of the linker and is directly attached to the 5’ end of the AIP. It is 18 amino acids (GGGGENLYFQGGKLGGGG) long and was designed to be efficiently cleaved by the TEV protease, as well as being codon-optimised for expression in B. subtilis. His-Tag: To be able to purify the protein for testing, we attached a His-Tag on our linker-AIP peptide. As it would probably interfere with recognition of the AIP by the receptor it has to be removed from the final construct. Stop Codon: In order to end translation a double stop codon was put in place.
Design: Pfu DNA polymerase was used to minimise PCR errors References: Very helpful if you have them! <biblio> </biblio> K316040 - sRBSName: Code: BBa_K316040 Sequence: Short: Synthetic RBS for LytC cell wall anchoring protein Long: Optimal Ribosome Binding Site (RBS) designed for high expression levels in B. subtilis. Opimised using Vogit Lab RBS calculator v 1.0 [http://voigtlab.ucsf.edu/software/] this software is based on
Design: Pfu DNA polymerase was used to minimise PCR errors References: Very helpful if you have them! |
| Parts |
 "
"