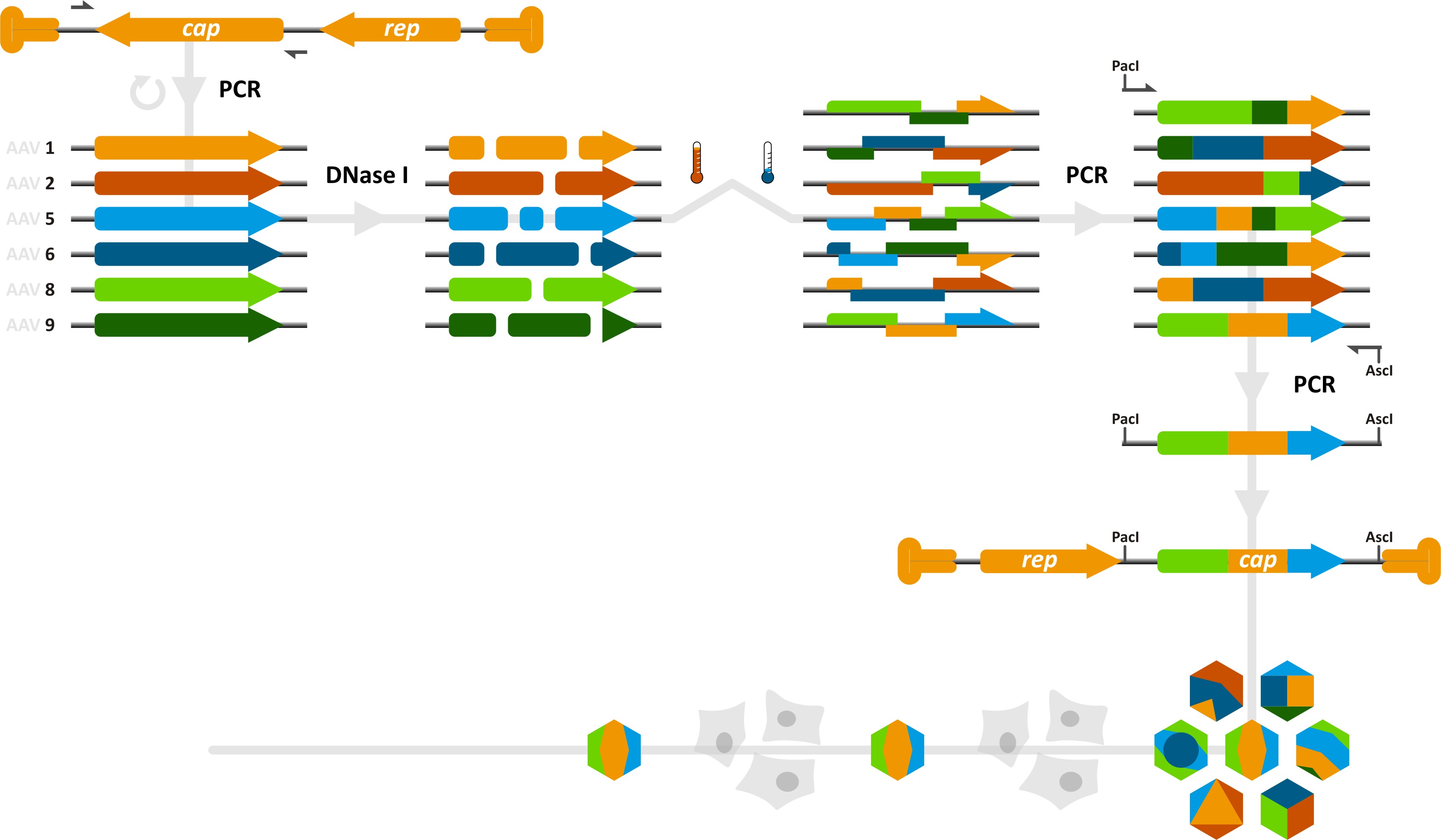
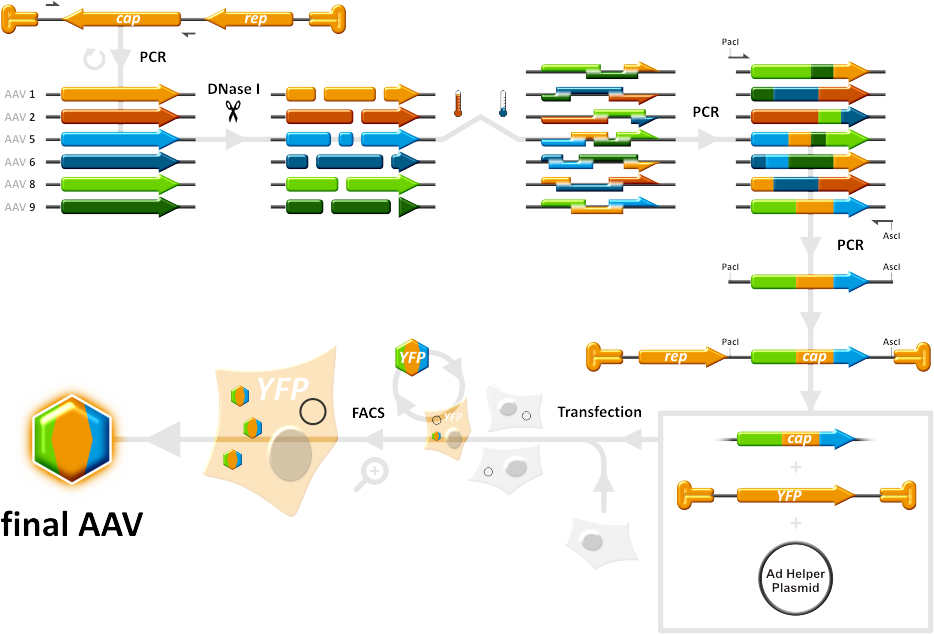
Team:Heidelberg/Project/Capsid Shuffling/Homology Based
From 2010.igem.org
| Line 26: | Line 26: | ||
{{:Team:Heidelberg/Pagemiddle}} | {{:Team:Heidelberg/Pagemiddle}} | ||
| + | |||
| + | [[Image:Shuffling sketch.jpg|270px]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
[[Image:HomBasShuffling 934px.png|270px]] | [[Image:HomBasShuffling 934px.png|270px]] | ||
{{:Team:Heidelberg/Bottom}} | {{:Team:Heidelberg/Bottom}} | ||
Revision as of 00:05, 28 October 2010

|
|
|||
 "
"