Team:Heidelberg/Notebook/miMeasure
From 2010.igem.org
(Difference between revisions)
(→Table2) |
(→miMeasure) |
||
| (23 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single}} |
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single_Pagetop|note_miMeasure}} |
| - | + | {{:Team:Heidelberg/Side_Top}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | {{:Team:Heidelberg/ | + | |
| - | + | ||
{| cellpadding="5" cellspacing="0" align="center" style="text-align: center; color:#4e93a4; border:1.53px solid #333333;" | {| cellpadding="5" cellspacing="0" align="center" style="text-align: center; color:#4e93a4; border:1.53px solid #333333;" | ||
|- border="0" | |- border="0" | ||
| Line 94: | Line 10: | ||
|colspan="3"| ||'''1'''||'''2'''||'''3'''||'''4''' | |colspan="3"| ||'''1'''||'''2'''||'''3'''||'''4''' | ||
|- style="background:#f2f2f2; color:#78b41e" | |- style="background:#f2f2f2; color:#78b41e" | ||
| - | |'''5'''||'''6'''||'''7'''||'''8'''||'''9'''||'''10'''||'''11''' | + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miMeasure/July#05.2F07.2F2010 5]'''||'''6'''||'''7'''||'''8'''||'''9'''||'''10'''||'''11''' |
|- style="background:#f2f2f2; color:#78b41e" | |- style="background:#f2f2f2; color:#78b41e" | ||
| - | |'''12'''||'''13'''||'''14'''||'''15'''||'''16'''||'''17'''||'''18''' | + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miMeasure/July#12.2F07.2F2010 12]'''||'''13'''||'''14'''||'''15'''||'''16'''||'''17'''||'''18''' |
|- style="background:#f2f2f2; color:#78b41e" | |- style="background:#f2f2f2; color:#78b41e" | ||
| - | |'''19'''||'''20'''||'''21'''||'''22'''||'''23'''||'''24'''||'''25''' | + | |'''19'''||'''20'''||'''21'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miMeasure/July#22.2F07.2F2010 22]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miMeasure/July#23.2F07.2F2010 23]'''||'''24'''||'''25''' |
|- style="background:#f2f2f2; color:#78b41e" | |- style="background:#f2f2f2; color:#78b41e" | ||
| - | |'''26'''||'''27'''||'''28'''||'''29'''||'''30'''||'''31'''||colspan="1"| | + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miMeasure/July#26.2F07.2F2010 26]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miMeasure/July#27.2F07.2F2010 27]'''||'''28'''||'''29'''||'''30'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miMeasure/July#31.2F07.2F20103 1]'''||colspan="1"| |
|- style="background:#f2f2f2; color:#78b41e" | |- style="background:#f2f2f2; color:#78b41e" | ||
|colspan="7"| | |colspan="7"| | ||
| + | <span style="color:#ffffff">-</span> | ||
|} | |} | ||
| Line 167: | Line 84: | ||
<span style="color:#ffffff">-</span> | <span style="color:#ffffff">-</span> | ||
|} | |} | ||
| + | |||
| + | {{:Team:Heidelberg/Side_Bottom}} | ||
| + | __NOTOC__ | ||
| + | =miMeasure= | ||
| + | |||
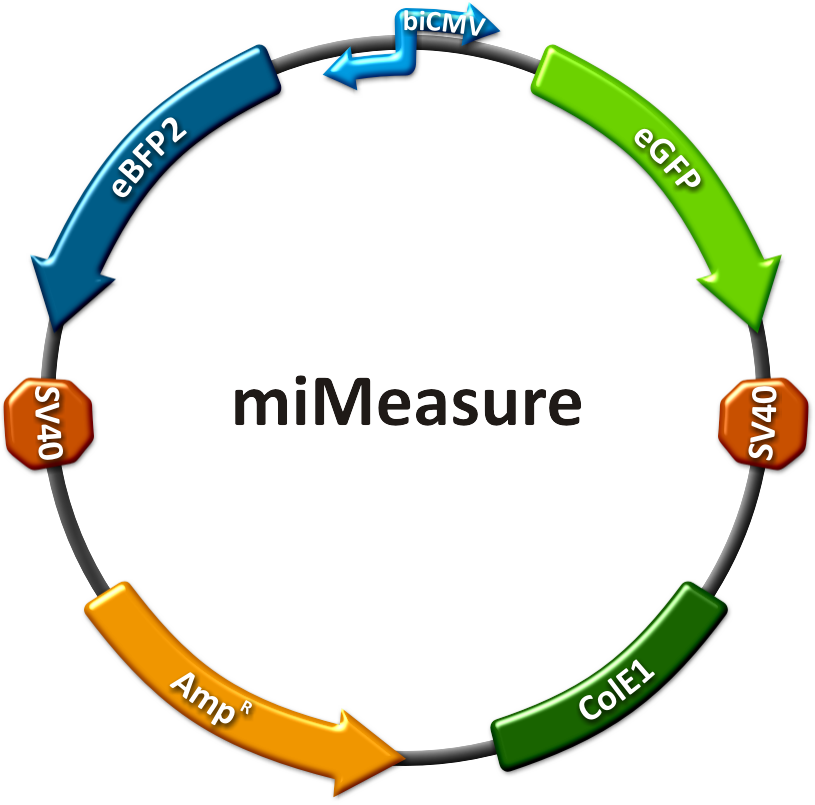
| + | miMeasure allows to quantitatively detect endogenous miRNA in different cells. To detect miRNA one should clone binding site for it into 3' UTR of GFP gene. To ensure that transfection efficiency or amount of cells do not affect result we decided to include reference fluorescence protein, BFP to allow normalisation. | ||
| + | |||
| + | |||
| + | [[Image:MiMeasure.png|500px|left|miMeasure plasmid]] | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single_Bottom}} |
| - | + | ||
Latest revision as of 03:53, 28 October 2010

miMeasuremiMeasure allows to quantitatively detect endogenous miRNA in different cells. To detect miRNA one should clone binding site for it into 3' UTR of GFP gene. To ensure that transfection efficiency or amount of cells do not affect result we decided to include reference fluorescence protein, BFP to allow normalisation.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"