Team:Heidelberg/Notebook/Homology Based/October
From 2010.igem.org
Laura Nadine (Talk | contribs) |
Philippbayer (Talk | contribs) (→22/10/2010) |
||
| (44 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single}} |
| + | {{:Team:Heidelberg/Single_Pagetop|note_homology}} | ||
| + | {{:Team:Heidelberg/Side_Top}} | ||
| + | {| cellpadding="5" cellspacing="0" align="center" style="text-align: center; color:#009be1; border: 1.5px solid #000000;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#f09600;" | [https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/August<font color="white">August</font>] | ||
| + | |- style="background:#f09600; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |colspan="6"| ||'''1''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''2'''||'''3'''||'''4'''||'''5'''||'''6'''||'''7'''||'''8''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''9'''||'''10'''||'''11'''||'''12'''||'''13'''||'''14'''||'''15''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''16'''||'''17'''||'''18'''||'''19'''||'''20'''||'''21'''||'''22''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''23'''||'''24'''||'''25'''||'''26'''||'''27'''||'''28'''||'''29''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''30'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/August#31.2F08.2F2010 31]'''||colspan="5"| | ||
| + | <span style="color:#ffffff">-</span> | ||
| + | |} | ||
| - | {{:Team:Heidelberg/ | + | |
| + | {| cellpadding="5" cellspacing="0" align="center" style="text-align: center; color:#78b41e; border: 1.5px solid #333333;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#009be1;" | [https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September<font color="#ffecba">September</font>] | ||
| + | |- style="background:#009be1; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |colspan="2"| ||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#01.2F09.2F2010 1]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#02.2F09.2F2010 2]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#03.2F09.2F2010 3]'''||'''4'''||'''5''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#06.2F09.2F2010 6]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#07.2F09.2F2010 7]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#08.2F09.2F2010 8]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#09.2F09.2F2010 9]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#10.2F09.2F2010 10]'''||'''11'''||'''12''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''13'''||'''14'''||'''15'''||'''16'''||'''17'''||'''18'''||'''19''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#20.2F09.2F2010 20]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#21.2F09.2F2010 21]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#22.2F09.2F2010 22]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#23.2F09.2F2010 23]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#24.2F09.2F2010 24]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#25.2F09.2F2010 25]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#26.2F09.2F2010 26]''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#27.2F09.2F2010 27]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#28.2F09.2F2010 28]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#29.2F09.2F2010 29]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/September#30.2F30.2F2010 30]'''||colspan="3"| | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |colspan="7"| | ||
| + | <span style="color:#ffffff">-</span> | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| cellpadding="5" cellspacing="0" align="center" style="text-align: center; color:#f09600; border: 1.5px solid #333333;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#78b41e;" | [https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October<font color="white">October</font>] | ||
| + | |- style="background:#78b41e; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |colspan="4"| ||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#01.2F10.2F2010 1]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#02.2F10.2F2010 2]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#03.2F10.2F2010 3]''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#04.2F10.2F2010 4]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#05.2F10.2F2010 5]''' | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#06.2F10.2F2010 6]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#07.2F10.2F2010 7]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#08.2F10.2F2010 8]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#09.2F10.2F2010 9]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#10.2F10.2F2010 10]''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#11.2F10.2F2010 11]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#12.2F10.2F2010 12]'''|'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#13.2F10.2F2010 13]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#14.2F10.2F2010 14]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/Homology_Based/October#15.2F10.2F2010 15]'''||'''16'''||'''17'''||'''18''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''19'''||'''20'''||'''21'''||'''22'''||'''23'''||'''24'''||'''25''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''26'''||'''27'''||'''28'''||'''29'''||'''30'''||colspan="3"| | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |colspan="7"| | ||
| + | <span style="color:#ffffff">-</span> | ||
| + | |} | ||
| + | |||
| + | {{:Team:Heidelberg/Side_Bottom}} | ||
| + | __NOTOC__ | ||
| + | =October= | ||
==01/10/2010== | ==01/10/2010== | ||
| Line 79: | Line 145: | ||
- A non-ITR AAV helper vector was obtained and maxi-preped to clone into it the cap genes from the 50 single clones picked. | - A non-ITR AAV helper vector was obtained and maxi-preped to clone into it the cap genes from the 50 single clones picked. | ||
| - | + | [[Image:102010_pEIKE_1-25.jpg|350px]] | |
| + | [[Image:102010_pEIKE_26-50.jpg|350px]] | ||
==09/10/2010== | ==09/10/2010== | ||
| Line 111: | Line 178: | ||
* The 6-well plate with primary hepatocytes was infected with the library AAVs, after 1 hour and a half MOI 10 and 100 Adeno-5 were added, starting the first selection round on these cells | * The 6-well plate with primary hepatocytes was infected with the library AAVs, after 1 hour and a half MOI 10 and 100 Adeno-5 were added, starting the first selection round on these cells | ||
| - | |||
| - | |||
==14/10/2010== | ==14/10/2010== | ||
| - | * 30 microliters of the supernatant collected | + | * Viruses from the 24-well-plate-Hek 293 cells from two days before were collected after monitoring the expression of YFP in those cells. Freeze-thaw cycles were done as previously, and the supernatant after centrifugation was collected. Each well represented one AAV. |
| + | |||
| + | * 30 microliters of the supernatant collected for each AAV were used to infect target cells, which were: | ||
- two 24-well plates HepG2 cells | - two 24-well plates HepG2 cells | ||
| Line 122: | Line 189: | ||
- Two 24-well plates Huh-7 cells | - Two 24-well plates Huh-7 cells | ||
| - | - Two 24-well plates primary hepatocytes | + | - Two 24-well plates primary hepatocytes |
| - | + | ||
| - | + | ||
==15/10/2010== | ==15/10/2010== | ||
| + | |||
| + | * AAVs from the second selection round on Huh-7 were collected from the supernatants after freeze-thaw cycles (see previous days), and stored at-20 C to infect the third selection round Huh-7 cells under the same conditions and in the same way as in the second selection round. | ||
* AAVs from the first selection round on primary hepatocytes were harvested using freeze-thaw cycles as before (see previous days), and the supernatants were kept at -20 C for the second selection round (primary hepatocytes are received every Monday). | * AAVs from the first selection round on primary hepatocytes were harvested using freeze-thaw cycles as before (see previous days), and the supernatants were kept at -20 C for the second selection round (primary hepatocytes are received every Monday). | ||
| + | ==16/10/2010== | ||
| + | * FACS measurements for the 6 24-well plates from two days earlier were done to measure YFP expression for the 48 different clones (AAVs!)in different kinds of cells. Cells were trypsinized (100 microliters trypsin) and then 500 microliters of buffer were added. Each well was measured for 1 min. The results showed no YFP expression for 47 of the samples in HepG2 and Huh-7, whereas one sample was positive (#43). This was confirmed further by microscopy. For the primary hepatocyes no sample showed any positive result. | ||
| - | + | Clone #43 was later injected into mice with a luc2-SV40 construct. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ==17/10/2010== | ||
| + | * Huh-7 cells were infected with AAVs in the supernatant from the second selection round, thus starting the third selection round. The same conditions and volumes as in the second selection round were used. | ||
| - | + | ==18/10/2010== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | * The single sample of AAV that showed a positive result for YFP measurement was sent for sequencing. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | * Primary hepatocytes were received and kept at 4 C for seeding the next day on 6-well plates. Collagen was used to coat the plates as before (Polymerization under UV for 1 hour was done). | ||
| - | + | ==19/10/2010== | |
| - | {{:Team:Heidelberg/ | + | |
| + | * Primary hepatocytes were seeded, and after 5 hours they were infected by AAVs from the first selection round. Adeno virus 5 MOI 100 was used, and varying volumes of the AAV-containing supernatant from the first selection round were applied. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==21/10/2010== | ||
| + | * Third selection round AAVs from Huh-7 and second selection round AAVs from primary hepatocytes were harvested by freeze-thaw cycles (see above). | ||
| + | |||
| + | * Viral DNA from the first, second and third selection rounds on Huh-7 and first and second selection rounds on primary hepatocytes was extracted using QIAmp MiniElute kit. | ||
| + | |||
| + | ==22/10/2010== | ||
| + | |||
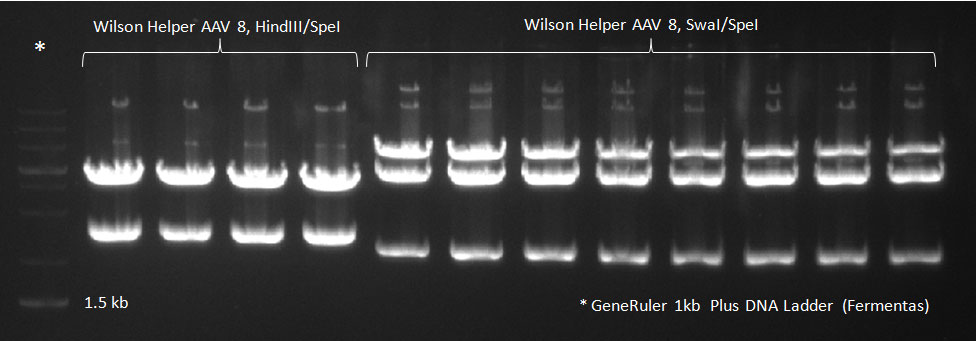
| + | * A PCR reaction using a primer mix was carried out on the DNA extracted the previous day (plus 20 of the clones picked earlier and clones into a non-ITR vector, see above) to amplify the cap genes and introduce SwaI and SpeI sites, which were then used to clone the cap genes into a Wilson wild-type vector. There was a problem with the non-ITR vector used earlier, in the sense that the expression of Cap and Rep genes was not comparable, so not many functional viral particles could be produced, and for this reason the Wilson wildtype vector was used instead. | ||
| + | The conditions for the PCR were the same as the second PCR for amplification of the shuffled capsids (see above). The bands for cap genes (2.2 KB) were purified from the gel, then digested with SwaI and SpeI, together with Wilson wild-type vector (standard 20 microliter digestion in NEB buffer 4, for 2 hours at 25 C). Qiagen nucleotide removal followed for the cap genes, and the vector was gel-purified. A ligation reaction of cap genes into the vector was carried out with NEB Quick Ligase (according to the standard manufacturer's recommendations), for 1 hour 30 min at room temperature. | ||
| + | |||
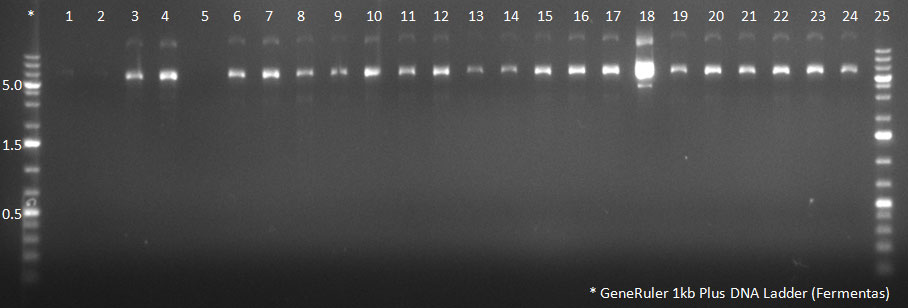
| + | [[Image:102310_20clones.gif|300px]] | ||
| + | PCR for Huh-7 clones | ||
| + | |||
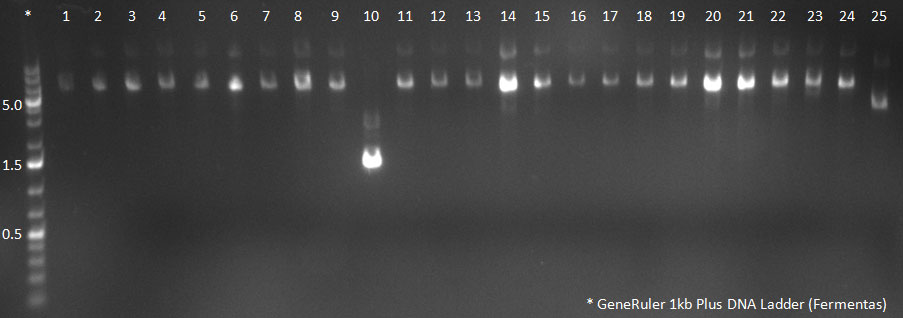
| + | [[Image:102310_H5-8_supern_PCR.jpg|300px]] | ||
| + | PCR for Huh-7 clones | ||
| + | |||
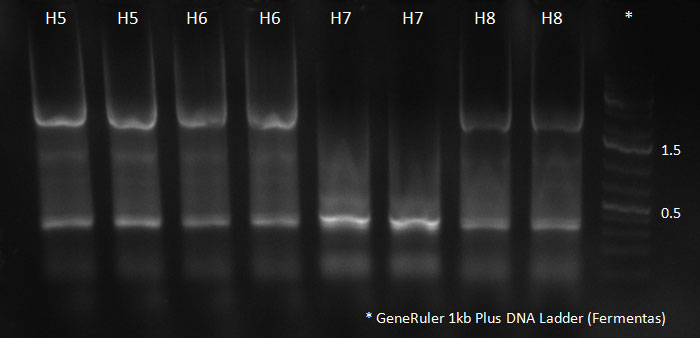
| + | [[Image:102310_H9-12_supern_PCR.jpg|300px]] | ||
| + | PCR for Huh-7 clones | ||
| + | |||
| + | [[Image:102310_Helper8_SpeI_HindIII.jpg|300px]] | ||
| + | Digested vector | ||
| + | |||
| + | [[Image:102310_Picked_Supern_PCR.jpg|300px]] | ||
| + | |||
| + | ==23/10/2010== | ||
| + | |||
| + | * The vector with cap genes from the previous day for the different selection rounds on Huh-7 and primary hepatocytes was transformed into Top10 cells. | ||
| + | |||
| + | ==24/10/2010== | ||
| + | |||
| + | * 20 clones were picked from each selection round on Huh-7 and primary hepatocytes from the transformation on the previous day and used to inoculate mini-preps. | ||
| + | |||
| + | ==25/10/2010== | ||
| + | |||
| + | * mini-preps were run for the samples from the previous day, and triple transfection of the Wilson construct with cap genes, Adeno-helper plasmid and a YFP construct with ITRs was done on 4 24-well plates of Hek293 cells in replica, each well containing an AAV construct from a different selection round on Huh-7 cells with different conditions during the selection process (washing after 30 min of AAV addition or no washing, see above). | ||
| + | |||
| + | From each selection round 12 samples were transfected, plus AAV wildtypes 1-12. | ||
| + | |||
| + | ==26/10/2010== | ||
| + | |||
| + | * The AAVs were harvested from the cells of one set of the plates (see previous day) using freeze-thaw cycles, and 20 microliters of the supernatant after centrifugation were used to infect HeLa, Huh-7 and primary hepatocyte cells in a 96-well plate format. This was done in replicates. The plates are then used for FACS analysis of YFP expression. | ||
| + | |||
| + | * The clones picked were sent for sequencing. | ||
| + | |||
| + | ==27/10/2010== | ||
| + | |||
| + | * A western blot and a dot blot for the other set of 24-well plates from two days earlier (see above) were initiated, with an antibody against a conserved region in cap genes. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | {{:Team:Heidelberg/Single_Bottom}} | ||
Latest revision as of 03:56, 28 October 2010

October01/10/2010
02/10/2010
03/10/2010
05/10/2010
- Washing the flasks with the medium they contain, and collecting the media from 13 flasks in 500 ml corning conical centrifuge tubes. - The cells were pelleted by centrifuging at 1500 rpm for 15 min at 4 ͦC - Discard supernatant - Washing: Resuspend the cells in 1X PBS and transfer to a 50 ml falcon - Pellet the cells again by centrifuging at 1500 rpm for 15 min at 4 ͦC - Discard supernatant
- Add 20 ml lysis buffer to the cell pellet - Put in liquid nitrogen for 5 min - Thaw at 37ͦC The cycles were repeated for 5 times, and the sample was frozen for next day. 06/10/2010
- The sample from previous day was sonicated in a sonication bath for 1 min 20 sec. - 50 units/ml benzonase were added to destroy RNA and DNA from non-AAV sources, which could later interfere with qPCR. - The sample was incubated at 37 ͦC for 30 min, and vortexed every 10 min. - The sample was then centrifuged for 15 min at 3270 xg - The supernatant was transferred into a new 50 ml falcon
- A Pasteur pipette was plugged into a Beckman quick-seal centrifuge tube - Using a 1000 µl pipette, 20 ml of the virus suspension were added the gradient was poured through the Pasteur pipette in the following order: - 7 ml 15% Iodixanol solution - 5 ml 25% Iodixanol solution - 4 ml 40% Iodixanol solution - 4 ml 60% Iodixanol solution - The Pasteur pipette was carefully removed and the tube was sealed. A tare tube was prepared in the same way. - Ultracentrifugation of the sample was carried out at 50,000 rpm for 2:30 hours at 4ͦC. - The virus-containing phase is the 40% iodixanol phase, which was sucked out using a syringe and a needle. The AAV sample was divided into aliquotes, one of them stored at 4 ͦC for qPCR the next day, and the rest were frozen in liquid nitrogen then transferred to -20 ͦC. 07/10/2010
- AAV MOI 100, 10 and 1 were used on Huh-7 cells plated on 6-well plates, and the viruses were allowed to infect cells for 1 hour and 30 min. - Adeno-virus 5 (MOI 10) was added then in a Biohazard II lab under the superviosion of lab personnel. - Cells were incubated in the Biohazard II lab. 08/10/2010- A non-ITR AAV helper vector was obtained and maxi-preped to clone into it the cap genes from the 50 single clones picked. 09/10/2010- The non-ITR vector was cut with AscI and PacI, and so was the pTR-UF3 containg cap genes from the 50 clones. The bands were purified from the gel.
10/10/2010
11/10/2010
13/10/2010
14/10/2010
- two 24-well plates HepG2 cells - Two 24-well plates Huh-7 cells - Two 24-well plates primary hepatocytes 15/10/2010
16/10/2010
Clone #43 was later injected into mice with a luc2-SV40 construct. 17/10/2010
18/10/2010
19/10/2010
21/10/2010
22/10/2010
The conditions for the PCR were the same as the second PCR for amplification of the shuffled capsids (see above). The bands for cap genes (2.2 KB) were purified from the gel, then digested with SwaI and SpeI, together with Wilson wild-type vector (standard 20 microliter digestion in NEB buffer 4, for 2 hours at 25 C). Qiagen nucleotide removal followed for the cap genes, and the vector was gel-purified. A ligation reaction of cap genes into the vector was carried out with NEB Quick Ligase (according to the standard manufacturer's recommendations), for 1 hour 30 min at room temperature. 300px PCR for Huh-7 clones 23/10/2010
24/10/2010
25/10/2010
From each selection round 12 samples were transfected, plus AAV wildtypes 1-12. 26/10/2010
27/10/2010
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"