Team:Freiburg Bioware/NoteBook/Labjournal/Cellculture
From 2010.igem.org
(→Fluorescence microscopy of transduction at 14.8 transfection of plates from 13.8) |
|||
| Line 984: | Line 984: | ||
<br /> | <br /> | ||
| + | ===<p style="font-size:17px; background-color:#00dd77;">XXXXXXXXXXXXXXXXXXXXXXXXXX </p>=== | ||
<h3>Harvesting viral stock from 18.8.2010 and following transduction</h3><br /> | <h3>Harvesting viral stock from 18.8.2010 and following transduction</h3><br /> | ||
<b>the motivation</b><br /> | <b>the motivation</b><br /> | ||
| Line 1,021: | Line 1,022: | ||
| - | + | ===<p style="font-size:17px; background-color:#00dd77;">XXXXXXXXXXXXXXXXXXXXXXXXXX </p>=== | |
<h3>Checking transduction efficiency via fluorescence microscopy</h3> | <h3>Checking transduction efficiency via fluorescence microscopy</h3> | ||
<b>the motivation</b><br /> | <b>the motivation</b><br /> | ||
| Line 1,054: | Line 1,055: | ||
<br /> | <br /> | ||
the optimal confluence of AAv293 cells is between 100 000 and 800 000 cells. This will be the field of investigation in the next experiments | the optimal confluence of AAv293 cells is between 100 000 and 800 000 cells. This will be the field of investigation in the next experiments | ||
| - | + | ===<p style="font-size:17px; background-color:#00dd77;">XXXXXXXXXXXXXXXXXXXXXXXXXX </p>=== | |
<h2>25.8: Transfection of AAV293 with different amounts of cells, and two plasmid concentrations</h2> | <h2>25.8: Transfection of AAV293 with different amounts of cells, and two plasmid concentrations</h2> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| Line 1,122: | Line 1,123: | ||
<br /> | <br /> | ||
<br /> | <br /> | ||
| + | ===<p style="font-size:17px; background-color:#00dd77;">XXXXXXXXXXXXXXXXXXXXXXXXXX </p>=== | ||
<h2>Seeding AAV293 for 10xTransfection</h2> | <h2>Seeding AAV293 for 10xTransfection</h2> | ||
<br /> | <br /> | ||
| Line 1,200: | Line 1,202: | ||
| - | + | ===<p style="font-size:17px; background-color:#00dd77;">XXXXXXXXXXXXXXXXXXXXXXXXXX </p>=== | |
<h2>28.8 Checking functionality of RC vectors </h2> | <h2>28.8 Checking functionality of RC vectors </h2> | ||
<br /> | <br /> | ||
| Line 1,258: | Line 1,260: | ||
| - | + | ===<p style="font-size:17px; background-color:#00dd77;">XXXXXXXXXXXXXXXXXXXXXXXXXX </p>=== | |
<h2>4.9 checking constructs (GOIs) with/without HGH and beta globin and reassembled as control</h2> | <h2>4.9 checking constructs (GOIs) with/without HGH and beta globin and reassembled as control</h2> | ||
| Line 1,302: | Line 1,304: | ||
<b></b> | <b></b> | ||
<br /> | <br /> | ||
| + | ===<p style="font-size:17px; background-color:#00dd77;">XXXXXXXXXXXXXXXXXXXXXXXXXX </p>=== | ||
<h2>6.9 checking five ganciclovir concentrations for two day incubation approach optimal medication</h2> | <h2>6.9 checking five ganciclovir concentrations for two day incubation approach optimal medication</h2> | ||
<br /> | <br /> | ||
| Line 1,342: | Line 1,345: | ||
[[Image:Freiburg10 CC 6.9 Checking Ganciclovir concentrations-CHART.jpg|800px]] | [[Image:Freiburg10 CC 6.9 Checking Ganciclovir concentrations-CHART.jpg|800px]] | ||
<br /> | <br /> | ||
| + | ===<p style="font-size:17px; background-color:#00dd77;">XXXXXXXXXXXXXXXXXXXXXXXXXX </p>=== | ||
<h2>22.9 Transfection with Volkers R/C constructs</h2> | <h2>22.9 Transfection with Volkers R/C constructs</h2> | ||
<br /> | <br /> | ||
| Line 1,375: | Line 1,379: | ||
<b>The results</b> | <b>The results</b> | ||
<font size="4" color="red">on the 23.9 it was shure that something went wrong with the nomenclature, so this approach is definitely not working => threwn away!</font> | <font size="4" color="red">on the 23.9 it was shure that something went wrong with the nomenclature, so this approach is definitely not working => threwn away!</font> | ||
| - | + | ===<p style="font-size:17px; background-color:#00dd77;">XXXXXXXXXXXXXXXXXXXXXXXXXX </p>=== | |
<br /><h2>22.9 Testing of Hannas 6 VP-constructs with different compositions (10%, 25%, 50% and 75% fractions) with a VP2-knockout construct</h2> | <br /><h2>22.9 Testing of Hannas 6 VP-constructs with different compositions (10%, 25%, 50% and 75% fractions) with a VP2-knockout construct</h2> | ||
<br /> | <br /> | ||
| Line 1,412: | Line 1,416: | ||
<br> [[Image:Freiburg 10 FACS Linker Libary.jpg|800px]] | <br> [[Image:Freiburg 10 FACS Linker Libary.jpg|800px]] | ||
<br /> | <br /> | ||
| + | ===<p style="font-size:17px; background-color:#00dd77;">XXXXXXXXXXXXXXXXXXXXXXXXXX </p>=== | ||
<h2>22.9 Testing pTERT_mVENUS in R/C P326 and P431</h2> | <h2>22.9 Testing pTERT_mVENUS in R/C P326 and P431</h2> | ||
<br /> | <br /> | ||
| Line 1,457: | Line 1,462: | ||
<br /> | <br /> | ||
| - | + | ===<p style="font-size:17px; background-color:#00dd77;">XXXXXXXXXXXXXXXXXXXXXXXXXX </p>=== | |
<h2>28.9 Testing Loop Insertions</h2> | <h2>28.9 Testing Loop Insertions</h2> | ||
<br /> | <br /> | ||
Revision as of 14:30, 9 October 2010
Team:Freiburg Bioware/SubNoteBook
- March (labday 1)
- April (labday 2 - 5)
- May (labday 6 - 17)
- June (labday 18 - 45)
- July (labday 46 - 75)
- August part 1 (labday 76 - 92)
- August part 2 (labday 93 - 106)
- September part 1 (labday 107 - 123)
- September part 2 (labday 124 - 135)
- October part 1 (labday 136 - 145 )
- October part 2 (labday 146 - 155 )
- October part 3 (labday 156 - 166 )
- November (labday 167 - 170 )
- Cellculture
Cellculture
This subpage is for cellculture, the structure of each experiment is following:
Date of first action, title of the experiment
The motivation
This short introduction provides an inside into our thoughts for this experiment.
The plan
In this section, the design of the experiment is explained.
The results
Interpretation and discussion of our results.
14.6 Seeding AAV293 for transfection
The motivation
We want to create viral stocks with the stratagene R/C, the gene of interest is mVenus. This is a qualitative control.
The plan
AAV 293 and HT1080 Cells have been splitted and plated out on 10 cm dishes for transfektion.
The Cells were accounted by using the Neubauer-Meteringchamber.
After harvesting by following the standard protocol the pallet have been resuspendiated in 15ml of DTT medium.
2,5µl of this cell suspension have been mixed with 47.5µl of trypan blue.
We counted 12,5x 10^6 cells/ml for the AAV293 cell line and 10x10^6 cells /ml for the HT1080 cell line.
The AAV 293 cells have been plated out on four dishes with 250µl of the cell suspension, on one plated with 1ml and on another dish with 1.5ml. note that that the date is wrong on the plates and the flasks! Cells have been already plated out on the 14th. of June.
We also seated two flasks one for each cell line. Therefor we used 1ml of the HT1080 cell suspension and two ml of the the AAV293 cells.
1)
- pAAV_iGEM_mVenus_YFP (glycerol stock B34, P39) P39 has the correct sequence (confirmed)
- pHelper
- pAAV_RC
2)
- pAAV_iGEM_mVenus_YFP (glycerol stock B33, P38) P38 (we dont know if P39 has the correct sequence!)was used because the amount of P39 was not sufficient enough for four Transfections!
- pHelper
- pAAV_RC
Plasmid concentrations:
pAAV_RC: 1 µg/µl
pHelper: 280 ng/µl
pAAV_iGEM_mVenus_YFP, P39: 180,83 ng/µl
pAAV_iGEM_mVenus_YFP, P38: 179,75 ng/µl
Cellculture clone 1 and 2 were transfected with P39. Cellculture clone 3 and 4 were transfected with P38.
We could only pipet 3,3 µg (instead of 10 µg) from each of the 3 plasmids into the 15 ml falcons due to insufficient amount of plasmid.
Adrian tried to examine the precipation of the CaCl2+ DNA Clusters. We have to optimize the pH of our 2xHBS at the moment it is 11.12.
The viral stocks are harvested according to standard protocol.
The results
According to the stratagene protocol, transduction efficencies about ~80% are achievable. We performed every step under guidance, so probably something is wrong with the buffers, cells or even the standard protocol. The next step is to repeat this transfection.
19.6 Seeding AAV293 for second transfection according to standard protocol
The motivation
We want to repeat the last transfection to exclude faults in practical
The plan
Six transfections were performed according to the standart protocol: Media:Freiburg10_Transfection_protocoll.pdf
Three 10 cm cellcluture dishes with 293 cells were transfected with 10 µg DNA (3,33 µg each plasmid) and the other three 10 cm cellculture dishes with 30 µg DNA (10 µg DNA each plasmid).
Plasmids:
- pAAV_iGEM-MCS_mVenus (P41), 507 ng/µl
- pAAV_RC , 1000 ng/µl
- pHelper , 280 ng/µl
- Transduction of 2x6 wells was successfully done (14.25)
- one six well was transduced with 10µg the other with 30µg DNA amount
The results
The Transduction efficency is still quite low! We have to optimize the stratagene protocol!
30.6 Examination of different Transduction methods
The motivation
We want to rise the amount of transduced particels, instead of 500µl, 1000µl of the AAV-stock getting pipetted into each well. the second plate gets stimulated via UV-light to boost the expression of DNA-Repair mechanisms. This should enhance the second strand synthesis of our ssDNA-GOI => a rise of YFP Expression!
The plan
1. Plate I(A is up)
| 3x10^5 cells | 3x10^5 cells | 3x10^5 cells |
| 3x10^5 cells | 3x10^5 cells | 3x10^5 cells |
Transduction:
| 1000 µl of Viral Stock DROPPED | 1000 µl of Viral Stock gently resuspended | 1500 µl of Viral Stock hard resuspending |
| 1000 µl of Viral Stock DROPPED | 1000 µl of Viral Stock gently resuspended | no Transduction |
2. Plate II (A is up)
| about 3x10^5 cells | only medium | only medium |
| about 3x10^5 cells | only medium | only medium |
Transduction:
| 1500 µl of Viral Stock gently resuspended | no Transduction | no Transduction |
| 1500 µl of Viral Stock gently resuspended | no Transduction | no Transduction |
The results
All cells are dead. It was not possible to take pictures because the dead cells were all clumped up.
Looks like we should not transduce 3x10^5 cells with 1000 µl (or more) of our viral solution because almost all cells died in all approaches and the medium indicator was yellow.
So we can not say if resuspendig the cells with the viral soultion is a useful alteration of the standard protocol.
3.7 Seeding AAV293 for transfection with different plasmid amounts and harvesting methods
The motivation
We want to check if the amount of plasmids is the reason for the low transduction efficiency. Our main problem is that the flow cytometry (FACS) is not available, so the results have to be checked via fuorescence microscopy.
The plan
Transfection with P41 was performed according standart protocol.
Two transfections where carried out with 10µg (3,33 µg each plasmid) DNA and the other two with 20µg DNA (6,66 µg each plasmid).
We want to investigate which of the following approaches yields a better transduction efficiency.
- Half of the transfected cells were exposed 3 cycles of thawing and freezing and then centrifuged (15 ml falcon, 2100G). The supernatant was transfered into a new 15 ml falcon and used for transduction.
- The other half was centrifuged at 300 G for 5min . The supernatant was transfered into another 15 ml falcon and the pellet was resuspended with 5 ml of DMEM. The content of these two falcons was also exposed to 3 cycles of thawing and freezing and then used for transduction.
- We have got 2 10 cm cellculture dishes with 10µg and 2 dishes with 20µg DNA used for transfection.
Approach with standart protocol (one dish with 10 and the other with 20µg Plasmids)(2100 G and 3 cycles of freezing and thawing)
Deviations from the standart protocol:
- The cells were centrifuged at 2100 G instead of 10.000 G
- The viruses were harvested after 44 hours
- 3 cycles of freezing and thawing
- used plasmids: 10µg, 20µg YFP
- we have 10µg and 20µg from standart protocol => SV
- we have 10µg and 20µg from standart protocol pellet => Pellet
- we have 10µg and 20µg from standart protocol suspension => Super
- amount of 500µl
1. Plate I(A is up)
| 10µg dropped Super | 10µg dropped Super gently resuspending | 10µg dropped Super gently resuspending |
| 10µg Super gently resuspending Super | 20µg dropped Super gently resuspending 1:10 dilution | control |
2. Plate I(A is up)
| 10µg dropped Super | 10µg dropped Super gently resuspending | 10µg dropped Super gently resuspending |
| 10µg Super gently resuspending Super | 20µg dropped Super gently resuspending 1:10 dilution | control |
3. Plate I(A is up)
| 10µg dropped Pellet | 10µg dropped Pellet gently resuspending | 10µg dropped Pellet gently resuspending |
| 10µg Super gently resuspending Pellet | 20µg dropped Pellet gently resuspending 1:10 dilution | control |
The results
We only took some pictures from single cells, because there was no significant difference in fluorescence between each sample.
The main conclusion of this experiment is, that we have to get the flow cytometry started.
4.7 Transfection with different amounts of plasmids
The motivation
We want to check if the mVenus expression/ transduction efficiency can be extended with higher amounts of DNA.
The plan
The Transfection was performed with 10, 18 and 24 µg per plasmid (mVenus, pHelper and R/C).
the harvest was performed according to standard protocol:
- 3 Plates got transduced
1. Plate I(A is up) completely 10 µg
| 300µl dropped | 300µl resuspended | 450µl resuspended |
| 600µl dropped | 600µl resuspended | control |
2. Plate II(A is up)completely 18 µg
| 300µl dropped | 300µl resuspended | 450µl resuspended |
| 600µl dropped | 600µl resuspended | control |
3. Plate III(A is up) completely 24 µg
| 300µl dropped | 300µl resuspended | 450µl resuspended |
| 600µl dropped | 600µl resuspended | control |
The results
FACS analysis of different treated HT1080 cells was done (18 samples)
The first percentage refers to the living and fluorescent cells and the second percentage to the total amout of living cells.
The data are listed this way:
1. Plate I(A is up)
| A1 | A2 | A3 |
| B1 | B2 | B3 |
1= Plate 1 A1 : 0% YFP-positive Cells (there were only a few cells in this sample)
2= Plate 1 A2 : 27,7% YFP-positve Cells from 73,3%
3= Plate 1 A3 : 17,3% " from 63,1%
4= Plate 2 A1 : 32,9% " from 75,3%
5= Plate 2 A2 : 27,2% " from 76,5%
6= Plate 2 A3 : 30,7% " from 73,4%
7= Plate 3 A1 : 26,8% " from 76,0%
8= Plate 3 A2 : 30,5% " from 74,2%
9= Plate 3 A3 : 30,7% " from 75,2%
10= Plate 1 B1 : 6,25% " from 36,1%
11= Plate 1 B2 : 6,42% " from 68,4%
12= Plate 1 B3 : 0,05% " from 89,5%
13= Plate 2 B1 : 20,5% " from 72,5%
14= Plate 2 B2 : 15,7% " from 73,6%
15= Plate 2 B3 : 0,02% " from 85,4%
16= Plate 3 B1 : 19,5% " from 73,3%
17= Plate 3 B2 : 20,8% " from 75,7%
18= Plate 3 B3 : 0,07% " from 84,8%
The calculated value written into the table describes the fluorescent amount of cells from the living cells. Example: 1 A2 : 27,7% YFP-positve cells of 73,3% living cells : 27,7/73,3 = 0,378
Conclusions
- Higher amounts of Plasmids (24, 30µg) seems to have no obvious effect on YFP expression
- Theres no difference in YFP expression comparing resuspended samples to not resuspended samples
- The amount of dead HT cells is about 25%, it is not clear if it is either AAV induced or mechanic stress (Trypsin, washing procedure)
Next steps:
- does the amount of GOI (ng plasmids) has an effect on YFP expression
- compare transduced to non transduced cells (vitality), or in other words is the mechanic stress responsible for 25% dead cell ratio?
- comparison of transduction efficiency of different virus stock production routines
14.7 Seeding AAV293 for TKGMK constructs
The motivation
We want to create the first viral particles with TKGMK as gene of interest.
The plan
30µg Plasmid were used (pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30)
VIRUS no GOI :pHelper + pAAV_RC
VIRUS 1 :pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30 clone 1 : P54
VIRUS 2 :pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30 clone 2 : P55
VIRUS 3 :pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30 clone 3 : P56
The results
Unfortunately the stocks never were used.
7.8 Seeding AAV293 for TKGMK stocks
11.8.2010 Transduction of HT1080 with no AAV, AAV without GOI and AAV with 10µg -> FACS
The motivation: In the past, the percentage of living cells in FACS was very low (~ 70%) so we want to check if the AAV-induced stress is responsible for cellular death, or the detaching procedure (trypsin and mechanical stress).
The plan
Plate I
| control, no cells | no GOI 150µl | 10µg 500µl |
| control, no virus | no GOI 300µl | 10µg 750µl |
Plate II
| control, no cells | no GOI 150µl | 10µg 500µl |
| control, no virus | no GOI 300µl | 10µg 750µl |
Plate III
| control, no cells | no GOI 150µl | 10µg 500µl |
| control, no virus | no virus | 10µg 750µl |
The results
ADD FACS DATA
The YFP expression is still quite low. This was kind of anticipated, the success in this experiment is the fact, that the amount of living cells is at a very high level! (~90%). The conclusion: Detach the cells fast! Use PBS which is not at 37°C (the PBS was 25min in the water bath), transfer the cells fast on ice after detaching!
The next steps are:
- Is it possible, that freeze/thaw circles reduce the transduction efficiency?
- Why is the YFP-expression that low? (~28-30%)
12.8 Virus production, seeding A431 cells
The motivation
In first place we want to create new stock of AAV without GOI, the next viral stock is with correct TK/GMK, the last stocks are with different YFP-amounts to check if the amount of GOI is a critical step for transduction effency (transgene expression).
The plan
See transduction protocoll for further information, the final cantrifugation step was done with 2000 g 5 min.
The results
Ten viral stocks with 3 ml each were prepared.
| Plate | pAAV_RC/µg | pHelper/µg | GOI/µg |
| 1 | 3,3 | 3,3 | no GOI |
| 2 | 6,6 | 3,3 | no GOI |
| 3 | 3,3 | 3,3 | 3,3 TK_GMK clone1 |
| 4 | 3,3 | 3,3 | 3,3 TK_GMK clone1 |
| 5 | 3,3 | 3,3 | 3,3 TK_GMK clone2 |
| 6 | 3,3 | 3,3 | 3,3 YFP |
| 7 | 3,3 | 3,3 | 10 YFP |
| 8 | 3,3 | 3,3 | 20 YFP |
| 9 | 3,3 | 3,3 | 40 YFP |
| 10 | 3,3 | 3,3 | 60 YFP |
The nomenclature of the plasmids is wrong, the date is 12.8. not 13.8
13.8 Seeding HT1080 for transduction at saturday, Seeding HEK293 for transfection
The motivation
The plan
Platte 1:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 1 | 300µl virus 2 |
| B | control, no virus | 600µl virus 1 | 600µl virus 2 |
Platte 2:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 3 | 300µl virus 4 |
| B | control, no virus | 600µl virus 3 | 600µl virus 4 |
Platte 3:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 5 | 300µl virus 6 |
| B | control, no virus | 600µl virus 5 | 600µl virus 6 |
Platte 4:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 7 | 300µl virus 8 |
| B | control, no virus | 600µl virus 7 | 600µl virus 8 |
Platte 5:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 9 | 300µl virus 10 |
| B | control, no virus | 600µl virus 9 | 600µl virus 10 |
Platte 6:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 3 | 300µl virus 4 |
| B | control, no virus | 600µl virus 3 | 600µl virus 4 |
Platte 7:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 3 | 300µl virus 4 |
| B | control, no virus | 500µl virus 3 | 500µl virus 4 |
Platte 8:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 5 | 300µl virus 5 |
| B | control, no virus | 600µl virus 5 | 600µl virus 5 |
The results
16.8: The fluorescence was quite low (~ 5%)in nearly every well! The 40µg YFP wells were the only exception.
The conclusions
The incubation step of CaCl2 seems to be crucial! The next transfection protocol is with a 30 min incubation on ice step!
Seeding HEK293 for transfection at monday 16.8
The motivation
Which confluence of HEK293 cells is optimal for AAV production?
The plan
We seed different amounts of cells, and check the highest titer.
| Plate | amount of cells |
| 1 | 100 000 |
| 2 | 200 000 |
| 3 | 400 000 |
| 4 | 500 000 |
| 5 | 800 000 |
| 6 | 1 000 000 |
| 7 | 1 200 000 |
| 8 | 1 500 000 |
| 9 | 1 750 000 |
| 10 | 1 750 000 not sure |
The results
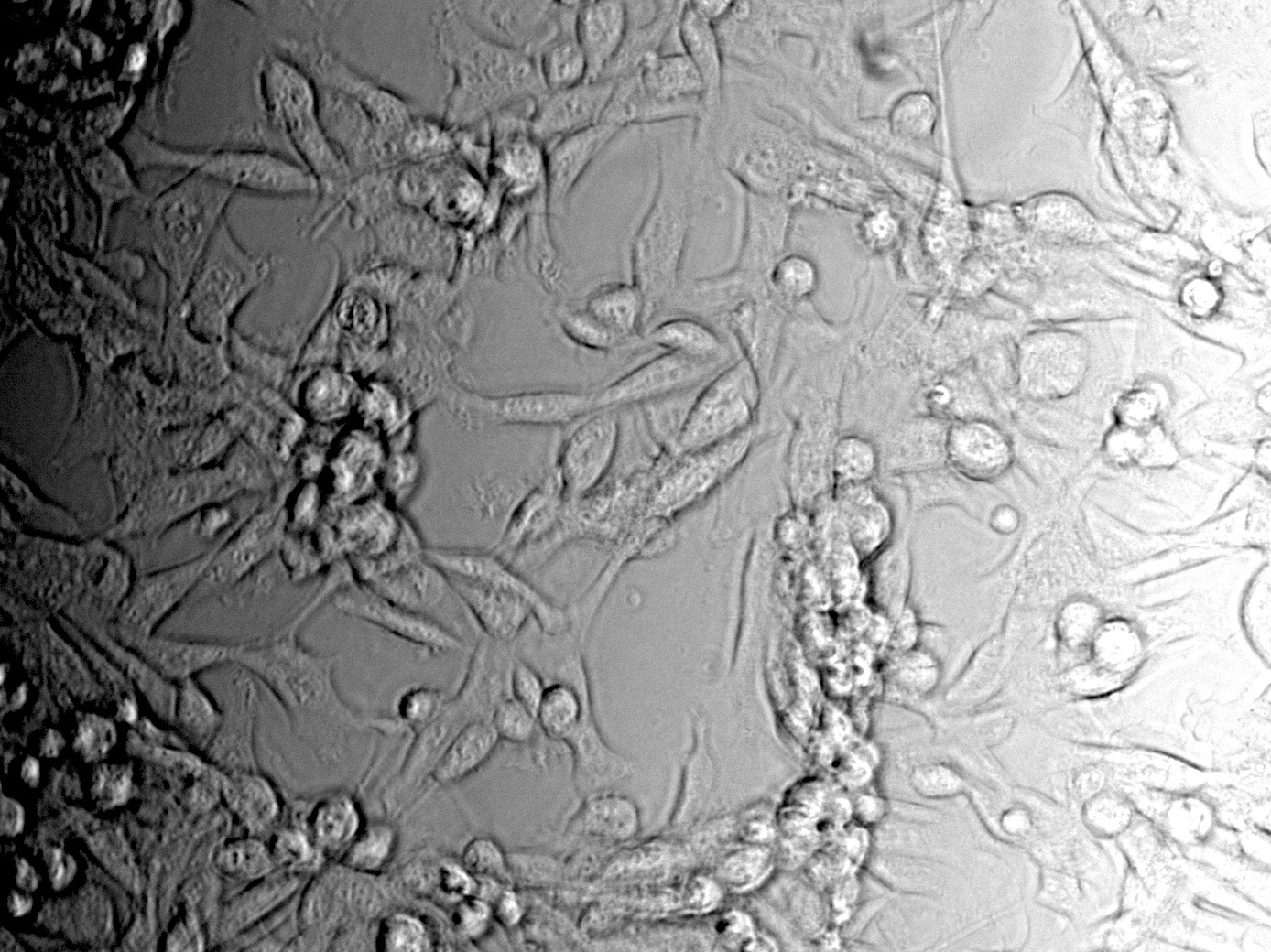
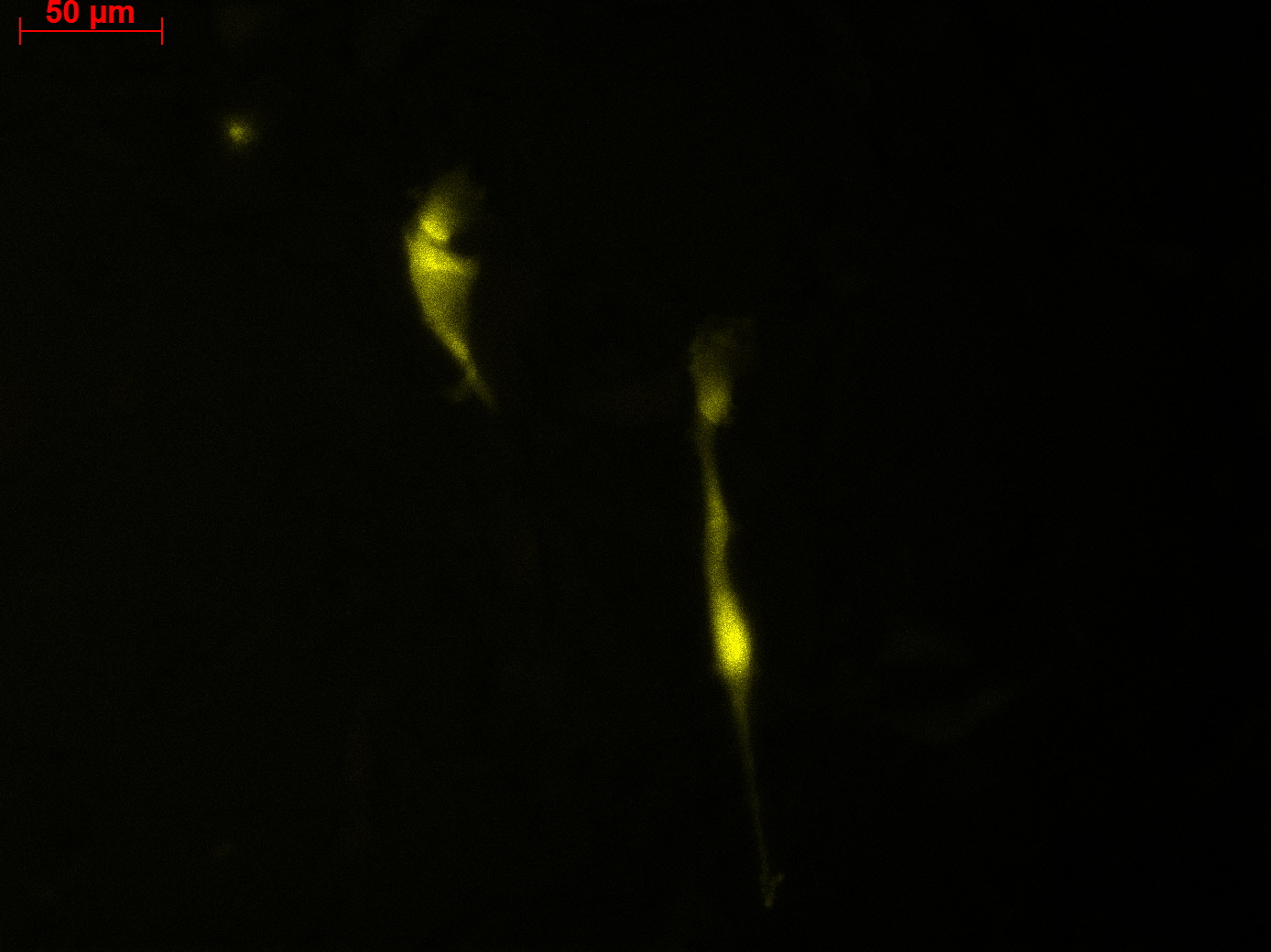
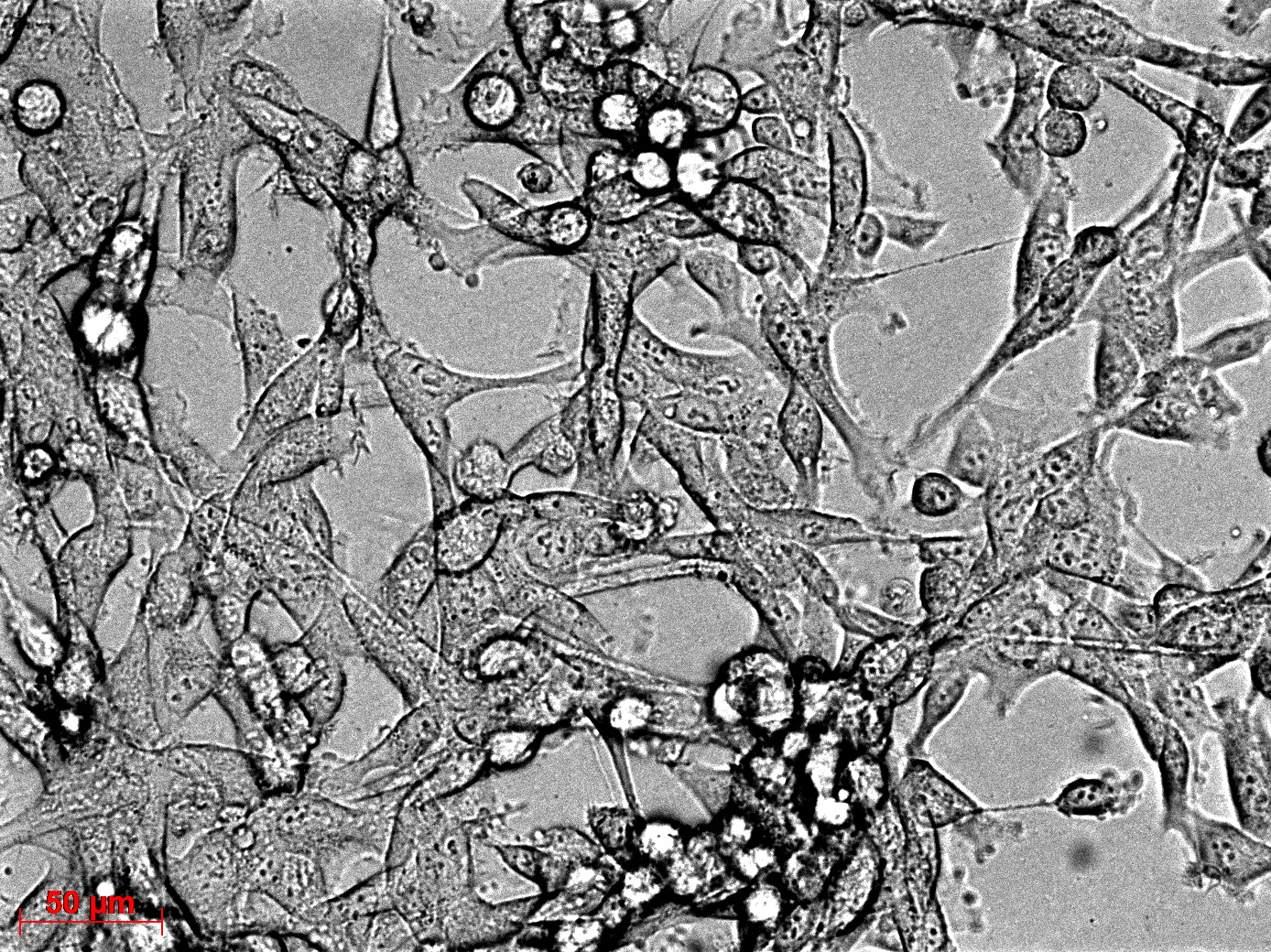
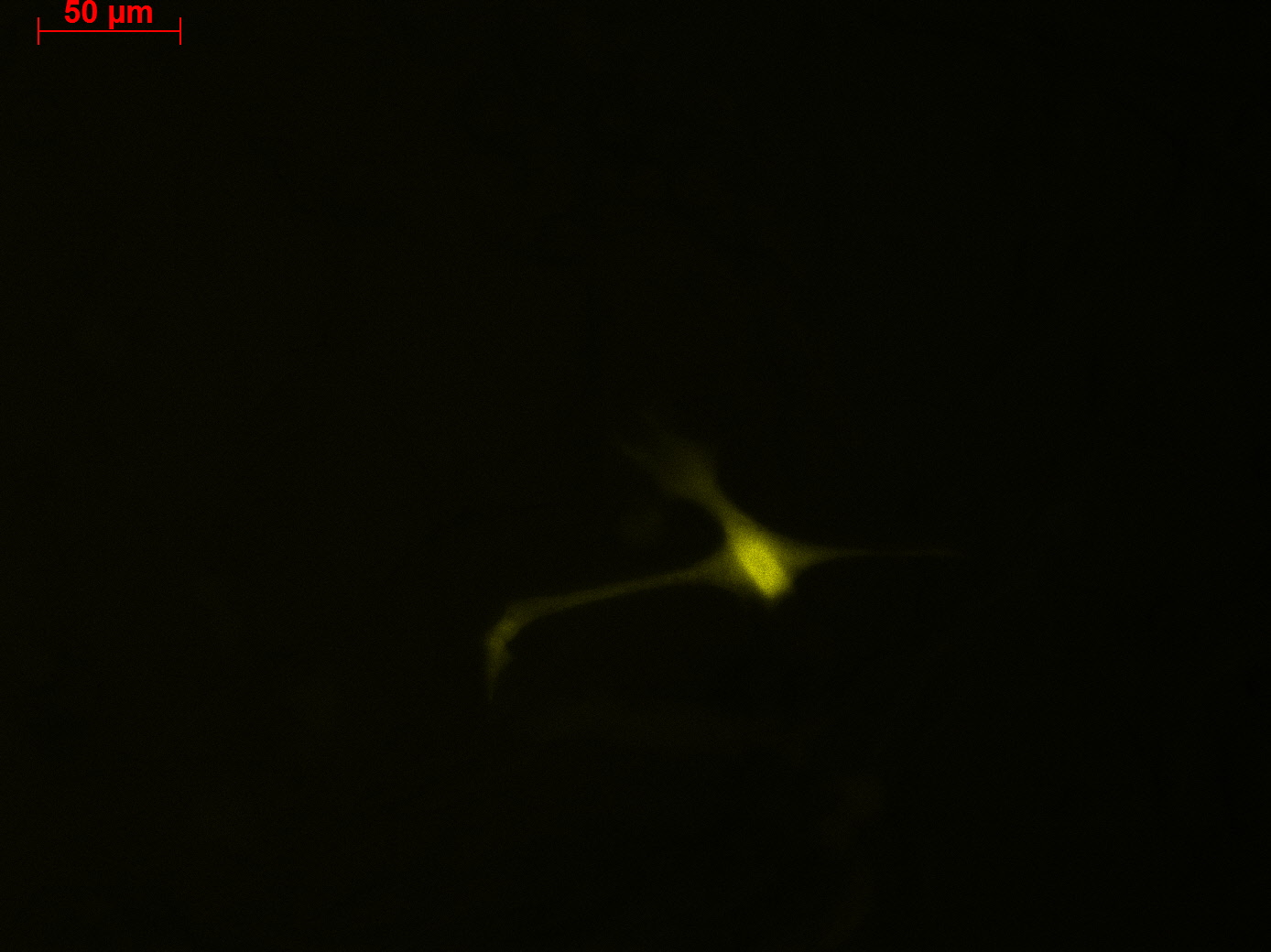
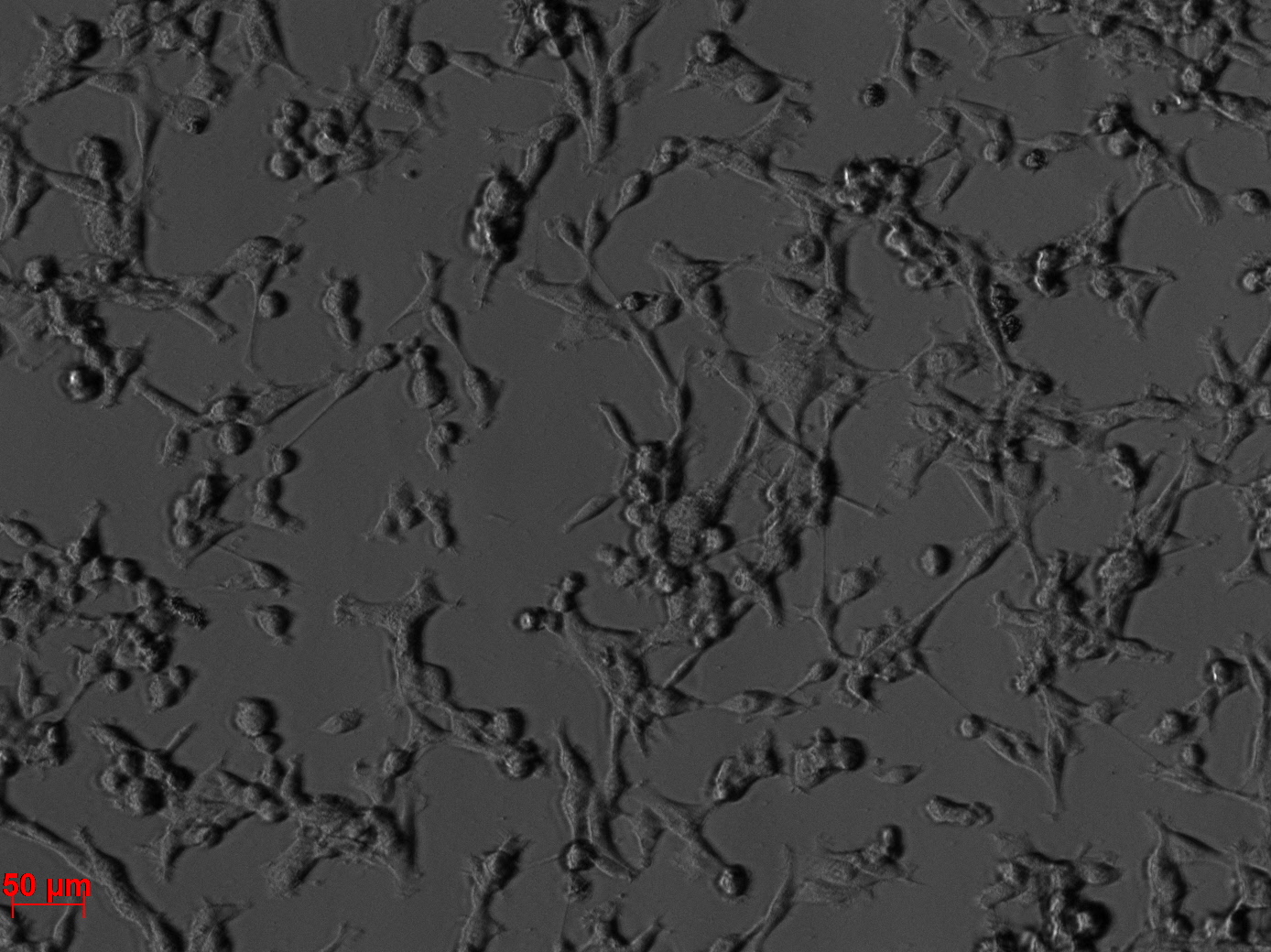
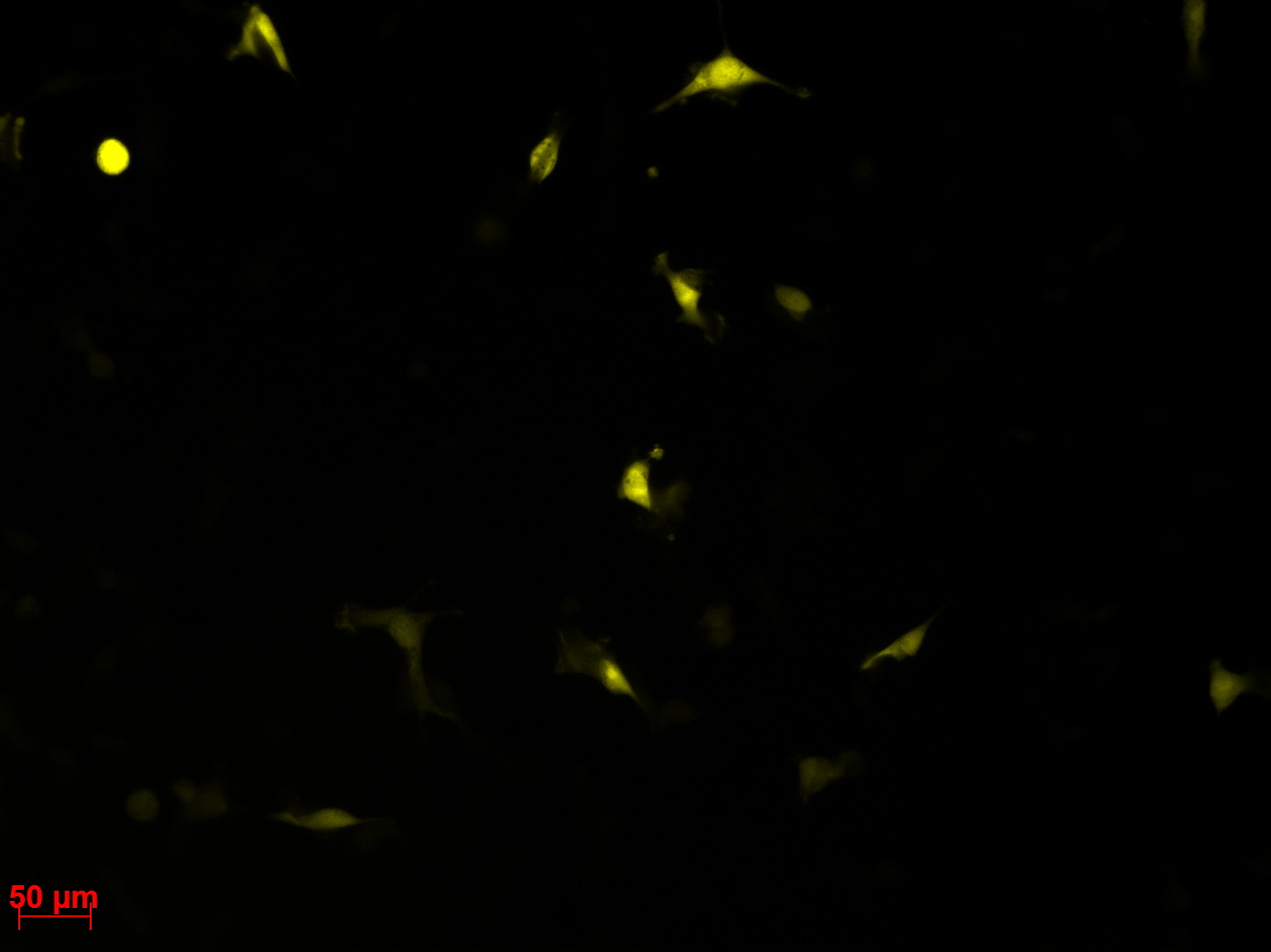
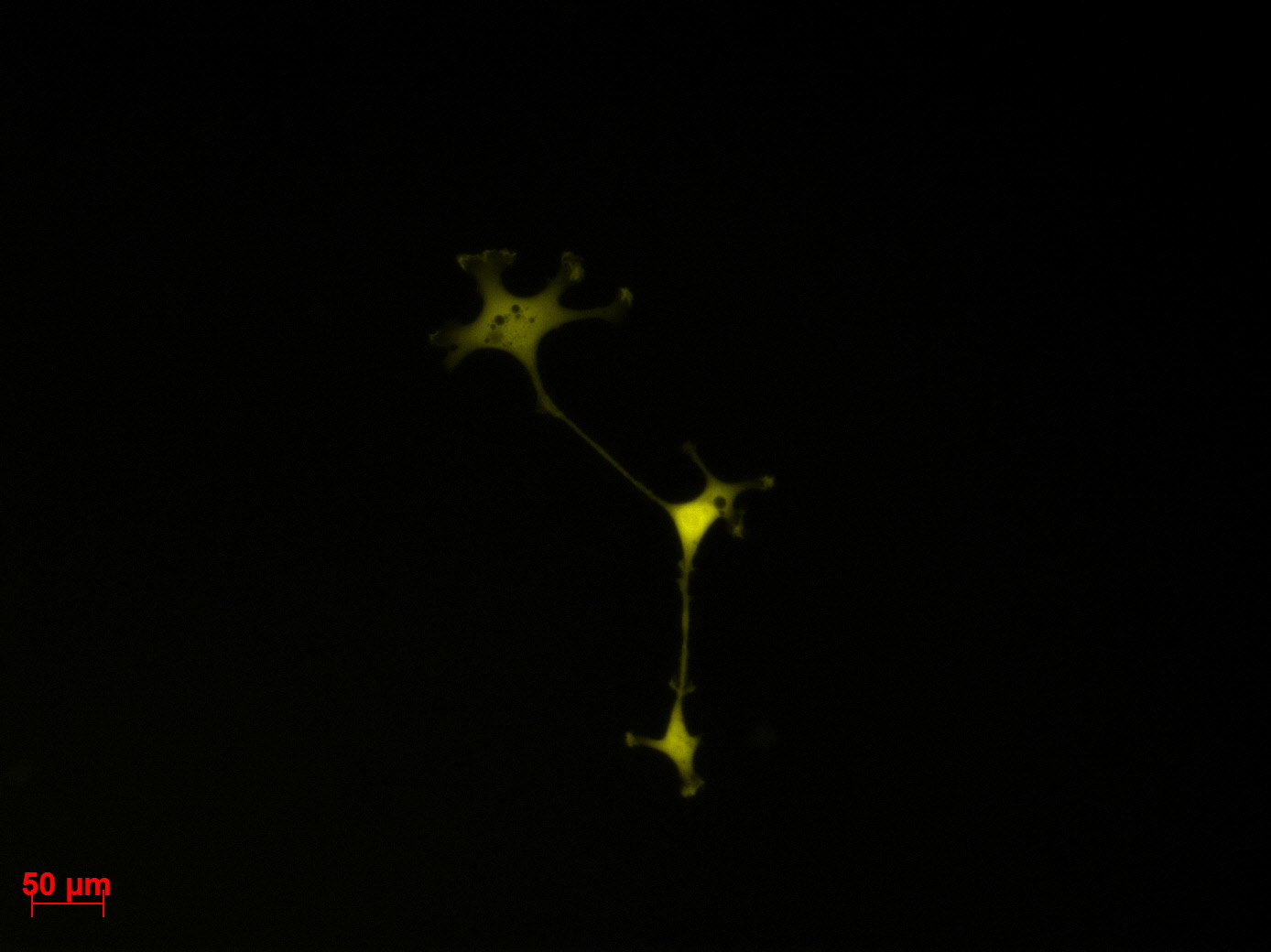
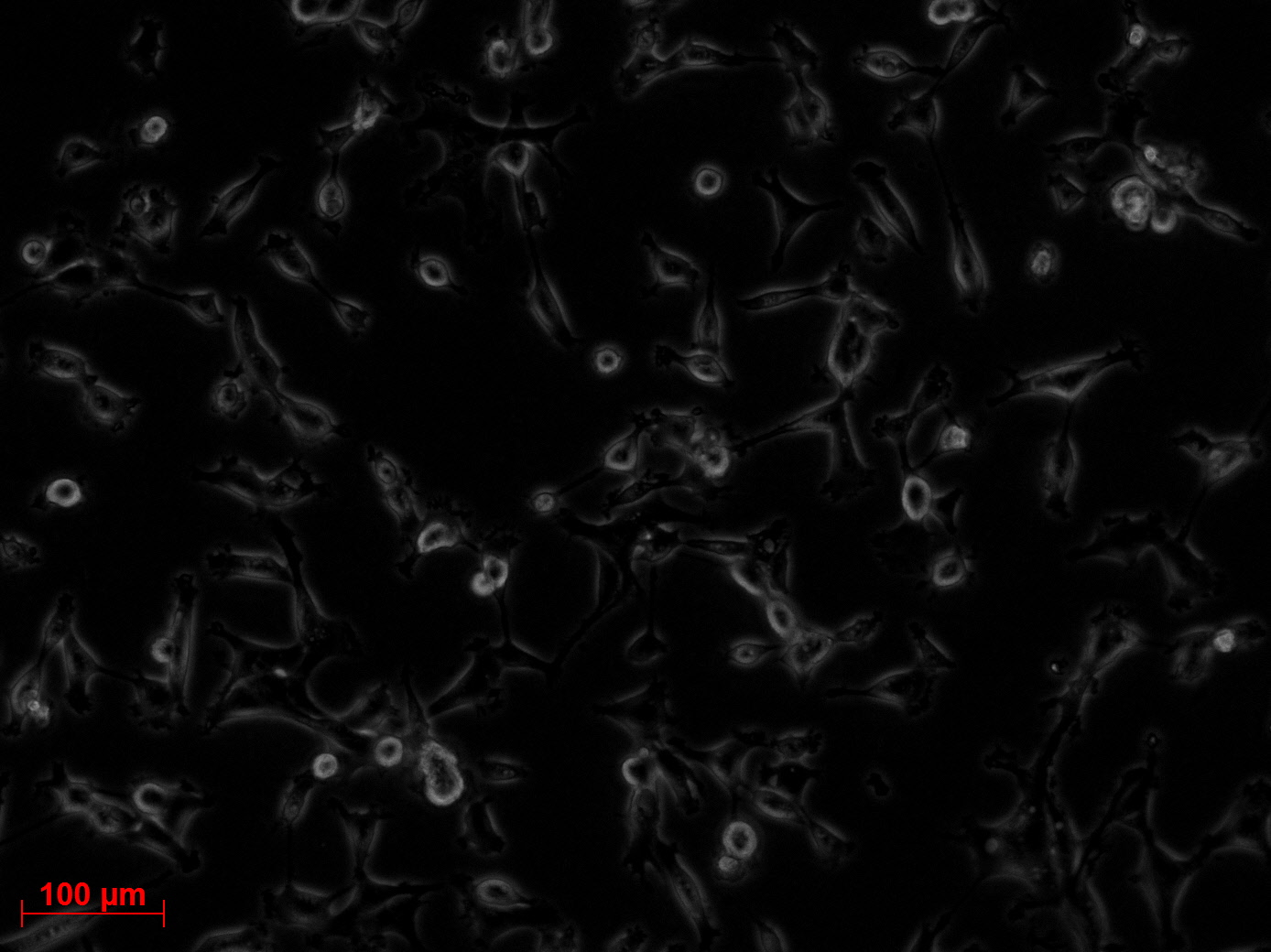
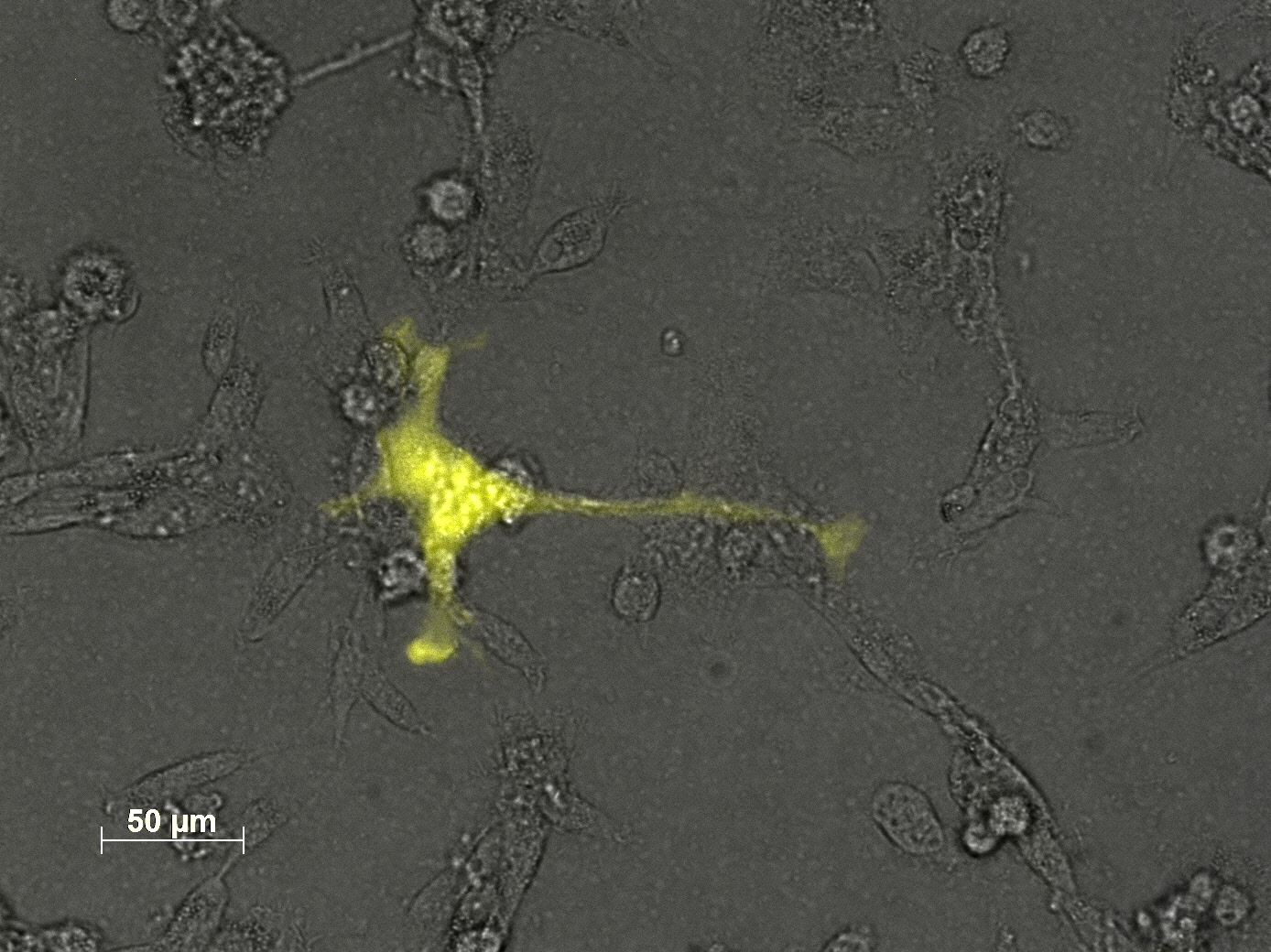
Fluorescence microscopy of transduction at 14.8 transfection of plates from 13.8
Working steps:
- Remove the three plasmids to be co-transfected (the recombinant pAAV expression plasmid or control plasmid, pAAV-RC, and pHelper) from storage at –20°C.
- Pipet XXX µg of each of the three plasmid DNA solutions into an eppi. The standart protocol is: 10 µg per plasmid, experiment data showed that the amounts (3,3 µg, 6,6 µg and 10 µg per plasmid) showed similar results
- Fill up to 300 µl with sterile water. Add 300µl of 0.5 M CaCl2 and mix gentlyIncubation step 45 min on ice !!!!!!
- Pipet 600 µl ml of 2 × HBS into a 15-ml falcon.
- Vortex the falcon (with the 2x HBS) gently!!!! while pipetting the DNA/CaCl2 solution dropwise into the falcon (use the Pipetus).
- Apply the DNA/CaCl2/HBS suspension to the plate of cells in a dropwise fashion(experimental data shows that resuspending-fashion is better for high titer), swirling gently to distribute the DNA suspension evenly in the medium.
- Return the tissue culture plate to the 37°C room for 6 hours.
- At the end of the incubation period, remove the medium from the plate and wash cells with warm PBS, replace it with 10 ml of fresh DMEM growth medium.This is a crucial step! The transfection-stress kills nearly all cells if theres no medium change
- Return the plate to the 37°C incubator for an additional 72 hours.
Seeding HT1080
The motivation
We want to check if the amount of HT cells is crucial for Transduction (The second transduction was daone with 30.000 cells, and had a high YFP expression)
The plan
3* 6er dishes with 100.000 cells and 2* 6er dishes with 50.000 cells were seeded.
The results
FILL IN PICTURES
Seeding AAV293
The motivation
We want to create new viral stocks.
The plan
15 * 10 cm2 with 2.000.000 cells on each plate were created
17.8 Transduction of plates with different amounts of cells
The plan
Platte 1 YFP: 50.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus | 300µl virus |
| B | control, no virus | 300µl virus | 300µl virus |
Platte 2 YFP: 50.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus | 300µl virus |
| B | control, no virus | 300µl virus | 300µl virus |
Platte 2 YFP: 50.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus | 300µl virus |
| B | control, no virus | 300µl virus | 300µl virus |
Platte 3 YFP: 100.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus | 300µl virus |
| B | control, no virus | 300µl virus | 300µl virus |
Platte 4 YFP: 100.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus | 300µl virus |
| B | control, no virus | 300µl virus | 300µl virus |
Platte 5 TKGMK: 100.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus | 300µl virus |
| B | control, no virus | 300µl virus | 300µl virus |
18.8 Transfection
The Motivation
Transfection with different amounts of REP/CAP and pHELPER. Additionally a
The plan
XXXXXXXXXXXXXXXXXXXXXXXXXX
Transfection of AAV293 cells
Investigator: Adrian, Kerstin
- 2x106 cells were seeded (instead of 3x106) --> hopefully better virus-production
- used plasmids:
- P50 c= 429 ng/µl (RC)
- P64 c= 500 ng/µl (pHelper)
- P229 c= 162 ng/µl (GOI=YFP)
- P228 c= 540 ng/µl (GOI=eGFP)
- P158a c= 1800ng/µl (RC mutant)
- P158a c= 1770ng/µl (RC mutant)
- 1. plate: Lipo-Transfection
- 2. plate: Lipo-Transfection
- 3. plate: 3,3 µg (RC), 3,3 µg (pHelper), 3,3 µg (YFP)
- 4. plate: 10 µg (RC), 10 µg (pHelper), 3,3 µg (YFP)
- 5. plate: 10 µg (RC mutant P158a), 10 µg (pHelper), 3,3 µg (YFP)
- 6. plate: 10 µg (RC mutant P158b), 10 µg (pHelper), 3,3 µg (YFP)
- 7. plate: 3,3 µg (RC mutant P158a), 3,3 µg (pHelper), 3,3 µg (YFP)
- 8. plate: 3,3 µg (RC mutant P158b), 3,3 µg (pHelper), 3,3 µg (YFP)
- 9. plate: 10 µg (RC), 10 µg (pHelper), 3,3 µg (eGFP)
- 10. plate: 10 µg (RC), 10 µg (pHelper), 10 µg (eGFP)
- 11. plate: 10 µg (RC), 10 µg (pHelper), 20 µg (eGFP)
- 12. plate: 10 µg (RC), 10 µg (pHelper), 40 µg (eGFP)
20.8
The Motivation
Seeding HT1080 for transfection with viral stocks from 18.8
The Plan
We harvest four T75 flasks and seed them into 6er dishes (200.000 cells in each well)
the Results
The Facs data at 22.8
21.8 Fluorescence microscopy, Transfection, AAV-Harvesting and Transduction
Transfection with different HBS-Buffers and with stratagene standard protocol
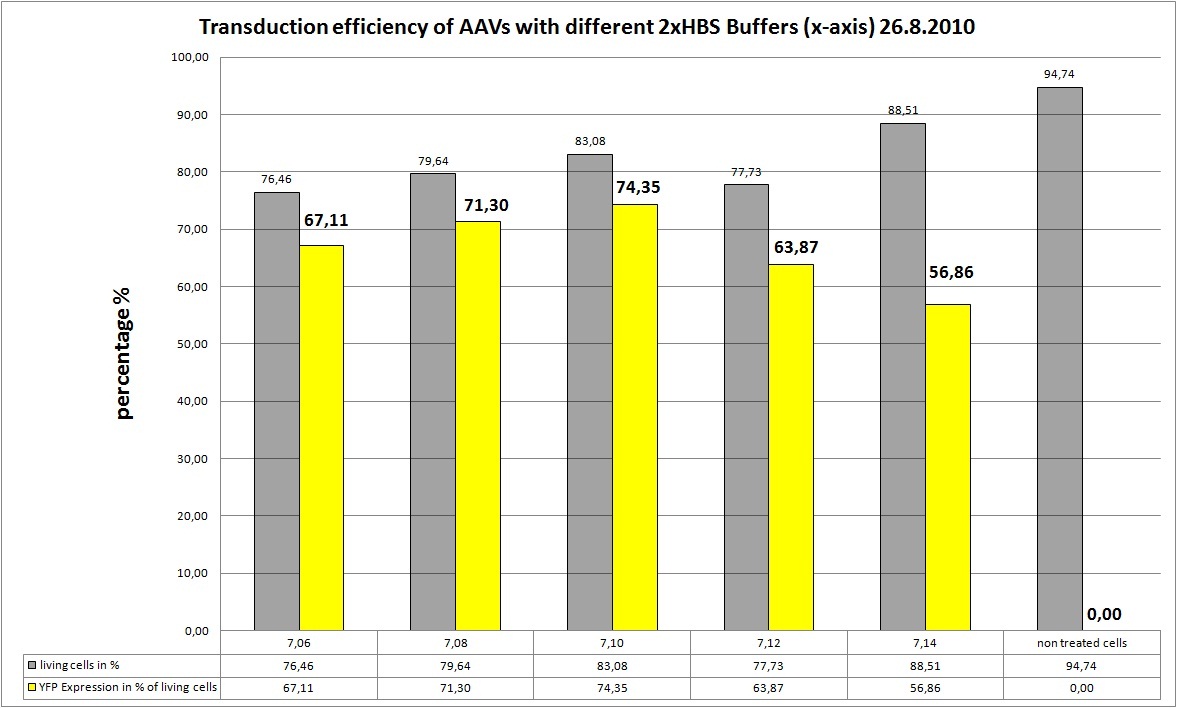
The Motivation
Transfection of AAV293 cells from 19.8 (ten 10cm2 dishes with 2.000.000 cells each) with original standard protocoll!!! We want to try different 2xHBS Buffers (pH) and their influence on transfection efficiency!
The Plan
Requiered goods: 0,5 M CaCl2 , 2x HBS-Buffer , autoclaved millipore water, falcons, eppis, sven's vortexer sterilized with EtOH
Working steps:
- Remove the three plasmids to be co-transfected (the recombinant pAAV expression plasmid or control plasmid, pAAV-RC, and pHelper) from storage at –20°C.
- Pipet 10 µg of each of the three plasmid DNA solutions into an eppi.
- Fill up to 300 µl with sterile water. Add 300µl of 0.5 M CaCl2 and mix gently
- Pipet 600 µl ml of 2 × HBS into a 15-ml falcon.
- Vortex the falcon (with the 2x HBS) gently!!!! while pipetting the DNA/CaCl2 solution dropwise into the falcon (use the Pipetus).
- Apply the DNA/CaCl2/HBS suspension to the plate of cells in a dropwise fashion, swirling gently to distribute the DNA suspension evenly in the medium.
- Return the tissue culture plate to the 37°C room for 6 hours.
- At the end of the incubation period, remove the medium from the plate and wash cells with warm PBS, replace it with 10 ml of fresh DMEM growth medium.
- Return the plate to the 37°C incubator for an additional 72 hours.
We try 5 different 2xHBS buffers:
- pH: 7.06
- pH: 7.08
- pH: 7.10
- pH: 7.12
- pH: 7.14
The transfected cells were seeded at 19.8 (2.000.000 cells per 10 cm2)
Used Plasmids: Standart protocol: 10µg of each plasmid
- GOI: mVENUS => P262, conc:1148ng/µl; used amount: 8,71µl
- GOI: TKGMK => P82b, conc:1500ng/µl; used amount: 6,67µl
- pHELPER => P64, conc:503ng/µl; used amount: 20µl
- RepCap => 10.8: 1348ng/µl; used amount: 7,41µl
- Remove the three plasmids to be co-transfected (the recombinant pAAV expression plasmid or control plasmid, pAAV-RC, and pHelper) from storage at –20°C.
- Pipet 10 µg of each of the three plasmid DNA solutions into an eppi.
- Add 1000µl of 0.3 M CaCl2 and mix gently
- Pipet 1000 µl ml of 2 × HBS into a 15-ml falcon.
- Pipett the DNA/CaCl2 solution dropwise into the falcon (use the Pipetus).
- Apply the DNA/CaCl2/HBS suspension to the plate of cells in a dropwise fashion, swirling gently to distribute the DNA suspension evenly in the medium.
- Return the tissue culture plate to the 37°C room for 6 hours.
- At the end of the incubation period, remove the medium from the plate and wash cells with warm PBS, replace it with 10 ml of fresh DMEM growth medium.
- Return the plate to the 37°C incubator for an additional 72 hours.
Eight new viral stocks were made with pHelper:20µl and RepCap:7,41µl each:
- mVenus:8,71µl; pH of 2xHBS:7,06
- mVenus:8,71µl; pH of 2xHBS:7,08
- mVenus:8,71µl; pH of 2xHBS:7,10
- mVenus:8,71µl; pH of 2xHBS:7,12
- mVenus:8,71µl; pH of 2xHBS:7,14
- TKGMK:6,67µl; pH of 2xHBS:7,08
- TKGMK:6,67µl; pH of 2xHBS:7,10
- TKGMK:6,67µl; pH of 2xHBS:7,12
the Results
AAV-Harvesting is at 24.8, same day Transduction and FACS will be done at 26.8
XXXXXXXXXXXXXXXXXXXXXXXXXX
Harvesting viral stock from 18.8.2010 and following transduction
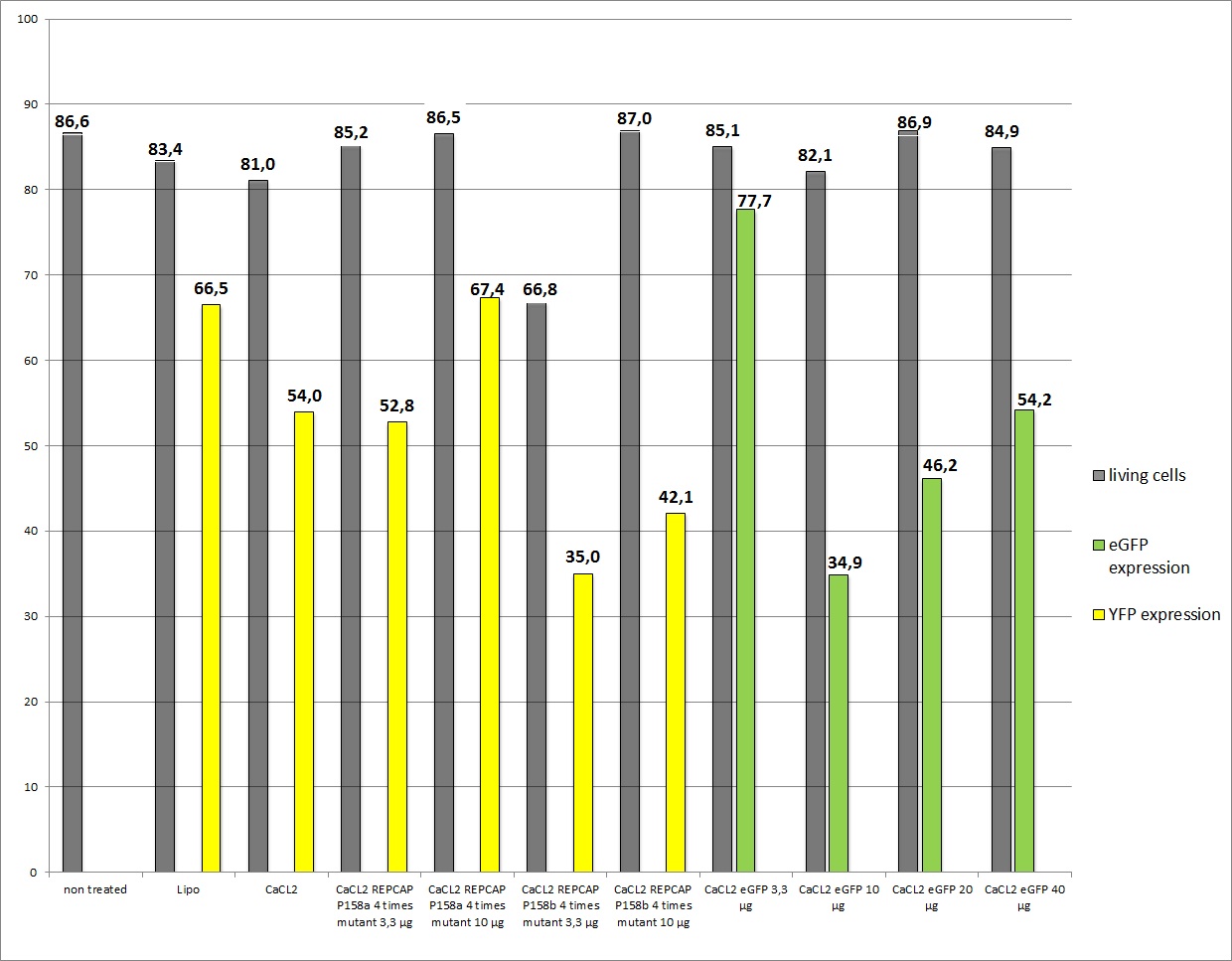
the motivation
We want to check the efficiency of Lipo-transfection and standard transfection with different ratios of REPCAP/pHelper, additionally if the modified RC (p158a and p158b) is still functionally.
The plan
Harvesting the cells with the Stratagene-standard protocoll.
The results
13 virals stocks were created (in 2.000.000 cells per 10cm2 dish):
- Lipo-Transfection 1 (2µl pHelper + 0,8µl RC + 6,2 µl YFP)
- Lipo-Transfection 2 (2µl pHelper + 0,8µl RC + 6,2 µl YFP)
- 3,3 µg of RC, pHelper and mVenus
- 10 µg RC, 10 µg pHelper, 3,3 µg GOI
- 10µg RC P158a four times mutant, 10µg pHelper, 3,3 µg mVenus
- 10µg RC P158b four times mutant, 10µg pHelper, 3,3 µg mVenus
- 3,3µg RC P158a four times mutant, 3,3µg pHelper, 3,3 µg mVenus
- 3,3µg RC P158b four times mutant, 3,3µg pHelper, 3,3 µg mVenus (negative control)
- 10µg RC, 10µg pHelper, 3,3µg GOI (broken arrow)
- 10µg RC, 10µg pHelper, 10µg GOI (broken arrow)
- 10µg RC, 10µg pHelper, 20µg GOI (broken arrow)
- 10µg RC, 10µg pHelper, 40µg GOI (broken arrow)
- 3,3 µg RC, 3,3 µg pHelper, 3,3 µg GOI
- negative control
- negative control
XXXXXXXXXXXXXXXXXXXXXXXXXX
Checking transduction efficiency via fluorescence microscopy
the motivation
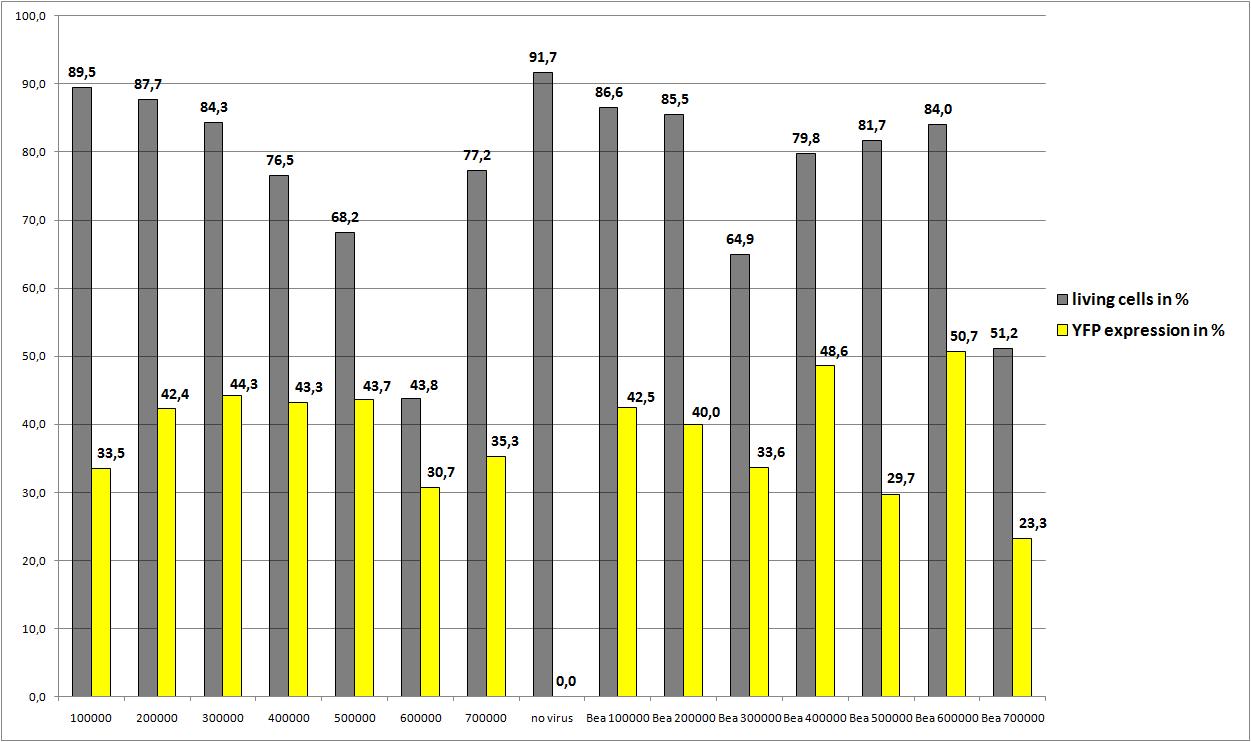
The transduction efficiency from the viral stocks which were created with different amounts of AAV293 was checked.
The plan
- plate 1:
- stock 1 (with 100.000 cells)
- stock 2 (with 200.000 cells)
- plate 2:
- stock 3 (with 400.000 cells)
- stock 4 (with 500.000 cells)
- plate 3:
- stock 5 (with 800.000 cells)
- stock 6 (with 1.000.000 cells)
- plate 4:
- stock 7 (with 1.200.000 cells)
- stock 8 (with 1.500.000 cells)
- plate 5:
- stock 9 (with 1.750.000 cells)
- stock 10 (with 1.750.000 cells)
The results
the optimal confluence of AAv293 cells is between 100 000 and 800 000 cells. This will be the field of investigation in the next experiments
XXXXXXXXXXXXXXXXXXXXXXXXXX
25.8: Transfection of AAV293 with different amounts of cells, and two plasmid concentrations
The motivation
We want to define the final AAV293-concentration for optimal AAV-Production
The plan
- 100.000
- 100.000
- 200.000
- 200.000
- 300.000
- 300.000
- 400.000
- 400.000
- 500.000
- 500.000
- 600.000
- 600.000
- 700.000
- 700.000
10µg RC P158a four times mutant, 10µg pHelper, 3,3 µg mVenus P162 were pipetted on plates: 2, 4, 6, 8, 10 , 12, 14
3,3µg RC P158a four times mutant, 3,3µg pHelper, 3,3 µg mVenus P262 were pipetted on plates: 1, 3, 5, 7, 9, 11, 13
| Days | 22.8 | 23.8 | 24.8 | 25.8 | 26.8 | 27.8 | 28.8 | 29.8 | 30.8 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
Data were not reliable (The amount of Goi (3,3 µg instead of 10µg) was not sufficient enough)so the average YFP expression is quite low, but the remarkable note is: Beas construct (pSB1C3_leftITR_CMV_betaglobin_mVenus_hGH_rightITR) fusioned from the created Biobricks still works!
XXXXXXXXXXXXXXXXXXXXXXXXXX
Seeding AAV293 for 10xTransfection
The motivation
We want to create ten hypothetical identical viral stocks to estimate the standard deviation.
The plan
| Days | 30.8 | 31.8 | 1.9 | 2.9 | 3.9 | 4.9 | 5.9 | 6.9 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | FACS |
Transfection will be done with:
- RepCap:P357 10µg =>conc.:1274,8ng/µl used amount: 7,84µl
- pHelper:P356 10µg =>conc.:1068ng/µl used amount: 9,36µl
- mVenus:P261 => excel sheet P263: 10µg =>conc.:979ng/µl used amount: 10,21 µl
0,5 ml of each stock become deepfreeze in.
One stock will be completely used for Transduction 9,5 ml (rest of the stock) * 0,5 ml (amount per transduction) => 19 values for estimating standard derivation.
getting valide data of 10µg samples
The motivation
we want to check the YFP expression
The plan
| Days | 30.8 | 31.8 | 1.9 | 2.9 |
|---|---|---|---|---|
| Action | seeding HT | Transduction | - | FACS |
One stock will be completely used for Transduction 9,5 ml (rest of the stock) * 0,5 ml (amount per transduction) => 19 values for estimating standard derivation.
the results
XXXXXXXXXXXXXXXXXXXXXXXXXX
28.8 Checking functionality of RC vectors
| Days | 28.8 | 29.8 | 30.8 | 31.9 | 1.9 | 2.9 | 3.9 | 4.9 | 5.9 | 6.9 |
|---|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | - | Transfection | - | - | Harvest+Seeding HT | Transduction | - | FACS |
Transfection was done with:
- RepCap:P326 => 2481 ng/µl used amount: 4µl => 10µg
- RepCap:P325 => 2051 ng/µl used amount: 4,87µl => 10µg
- pHelper:P356 => 684 ng/µl used amount: 414,62µl => 10µg
- mVenus:P262 => 1148 ng/µl used amount: 8,7µl => 10µg
The results
Transfection will be done with:
- RepCap:P357 10µg =>conc.:1274,8ng/µl used amount: 7,84µl
- pHelper:P356 10µg =>conc.:1068ng/µl used amount: 9,36µl
- mVenus:P261 => excel sheet P263: 10µg =>conc.:979ng/µl used amount: 10,21 µl
XXXXXXXXXXXXXXXXXXXXXXXXXX
4.9 checking constructs (GOIs) with/without HGH and beta globin and reassembled as control
The motivation
We want to check if these reassmbled constructs (GOIs) still work!
- P269 lITR_CMV_beta-globin_mVenus_HGH_rITR = 325 ng/µl => 9,66 µl
- P270 lITR_CMV_beta-globin_mVenus_HGH_rITR = 561 ng/µl => 7,26 µl
- P377 lITR_CMV_mVenus_HGH_rITR = 325,04 ng/µl => 30,76 µl
- P378 lITR_CMV_beta-globin_mVenus_rITR = 561 ng/µl => 17,8µl
All constructs are completed with p50 R/C =>> 26,45 µl and pHELPER 14,61µl
The plan
| Days | 4.9 | 6.9 | 7.9 | 8.9 | 9.9 | 10.9 | 11.9 | 12.9 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | FACS |
XXXXXXXXXXXXXXXXXXXXXXXXXX
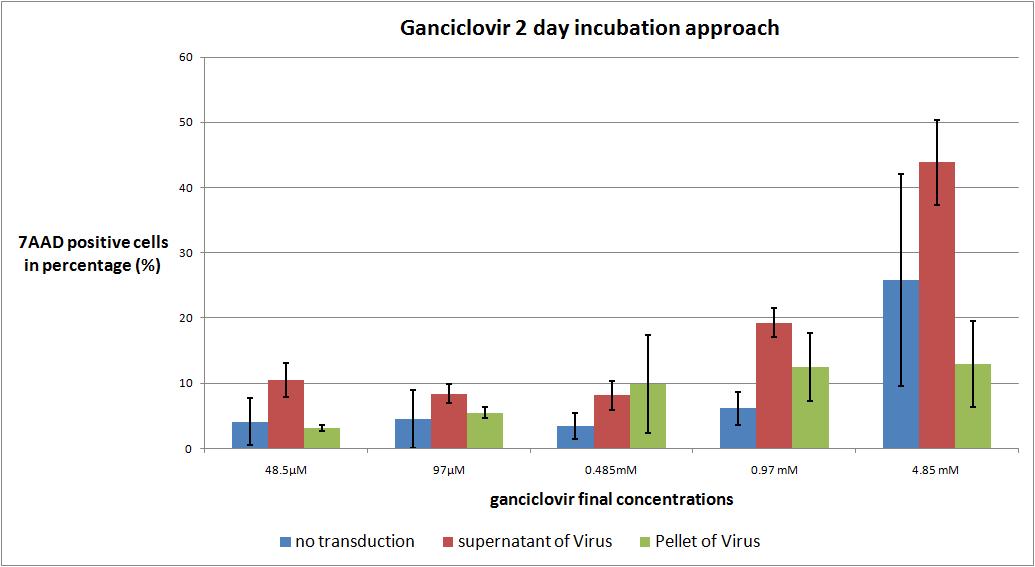
6.9 checking five ganciclovir concentrations for two day incubation approach optimal medication
The motivation
we want to check the optimal ganciclovir concentration, we use the TK/GMK Stocks (10 µg from each plasmid) from 24.8 with Buffer pH 7,10 and the pH12 stock.
The plan
| Days | 3.9 | 4.9 | 5.9 | 6.9 |
|---|---|---|---|---|
| Action | seeding HT | Transduction | - | FACS |
We want to use five different ganciclovir concentrations (the concentrations are absolute to the volume in the wells):
- 48.5µM
- 97 µM
- 0.485 mM
- 0.97 mM
- 4.85 mM
XXXXXXXXXXXXXXXXXXXXXXXXXX
22.9 Transfection with Volkers R/C constructs
The motivation
We want to check if these R/C still work!
The plan
| Days | 20.9 | 21.9 | 22.9 | 23.9 | 24.9 | 25.9 | 26.9 | 27.9 | 28.9 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293: 4*6wells | - | Transfection: 4 constructs | - | - | Seeding HT | Harvest + Transduction | - | FACS |
The results on the 23.9 it was shure that something went wrong with the nomenclature, so this approach is definitely not working => threwn away!
XXXXXXXXXXXXXXXXXXXXXXXXXX
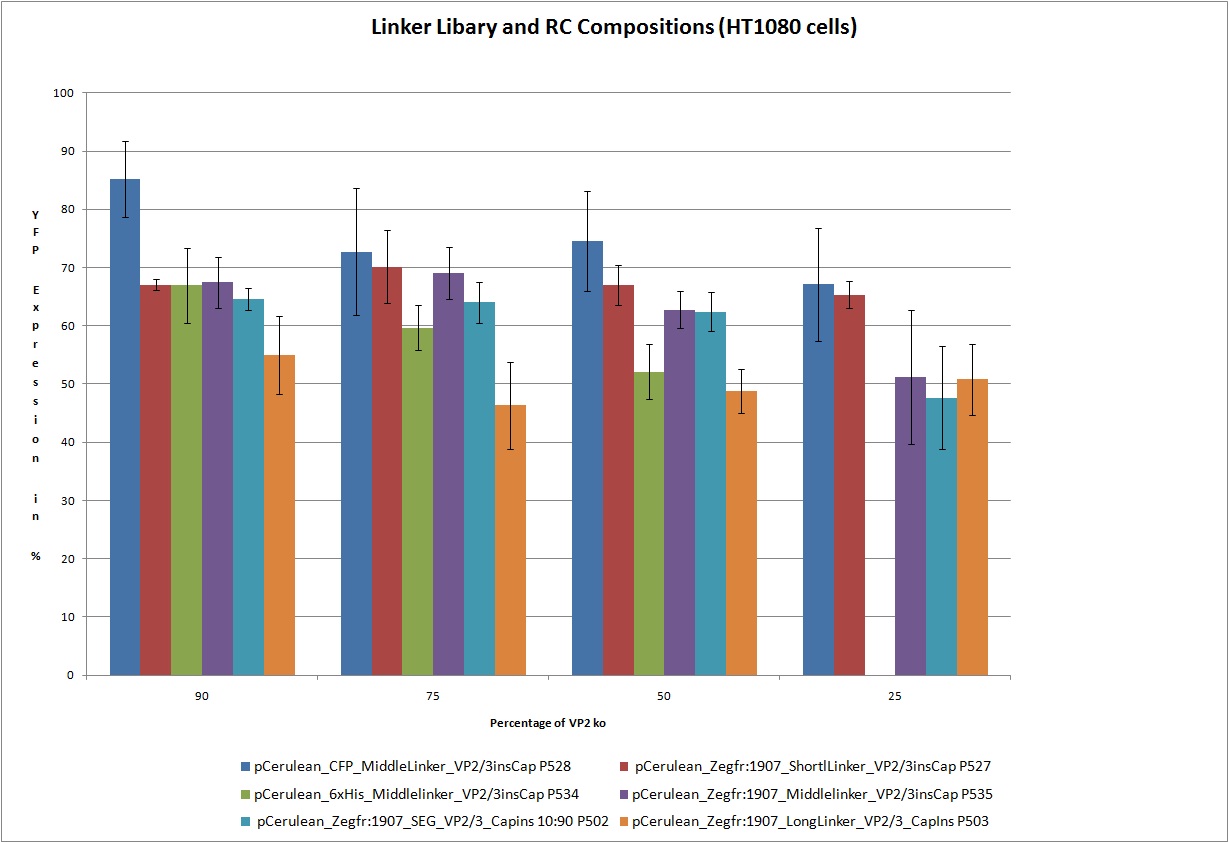
22.9 Testing of Hannas 6 VP-constructs with different compositions (10%, 25%, 50% and 75% fractions) with a VP2-knockout construct
The motivation
Test the 18 constructs for functionality on A431 and HT1080 cells.
The plan
| Days | 20.9 | 21.9 | 22.9 | 23.9 | 24.9 | 25.9 | 26.9 | 27.9 | 28.9 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
XXXXXXXXXXXXXXXXXXXXXXXXXX
22.9 Testing pTERT_mVENUS in R/C P326 and P431
The motivation
We want to check if the pTERT-promoter is realiable for tumor specific gene expression.
The plan
| Days | 22.9 | 23.9 | 24.9 | 25.9 | 26.9 | 27.9 | 28.9 | 29.9 | 30.9 | 1.10 | 2.10 | 3.10 | 4.10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest | - | - | Seeding HT, A293 and A431 | Transduction | FACS |
The results
XXXXXXXXXXXXXXXXXXXXXXXXXX
28.9 Testing Loop Insertions
The motivation
The plan
| Days | 28.9 | 29.9 | 30.9 | 1.10 | 2.10 | 3.10 | 4.10 | 5.10 | 6.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
The results
Experiments
The motivation
The plan
| Days | 22.9 | 23.9 | 24.9 | 25.9 | 26.9 | 27.9 | 28.9 | 29.9 | 30.9 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
The results
Citations
[1] Media:Freiburg10_Stratagene_AAV_helper_free_manual.pdf
[2] [http://www.copewithcytokines.de/cope.cgi?key=A431 Titel einfügen]
[3] Read, A. P.; Strachan, T. (1999). "Chapter 18: Cancer Genetics". Human molecular genetics 2. New York: Wiley. ISBN 0-471-33061-2.
[4] [http://www.biomol.de/dateien/infos_nr778.gif Titel einfügen]
[5] Media:Freiburg10 Titration of AAV-2 particles via a novel capsid ELISA.pdf
 "
"