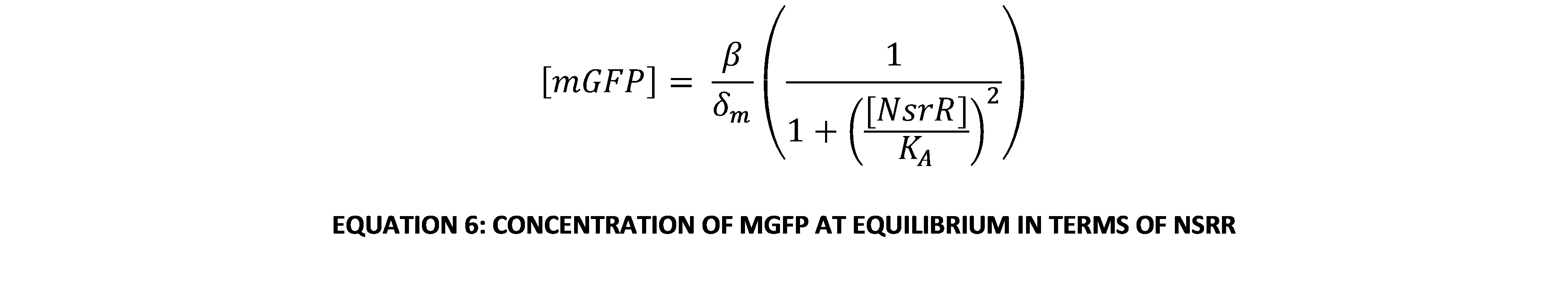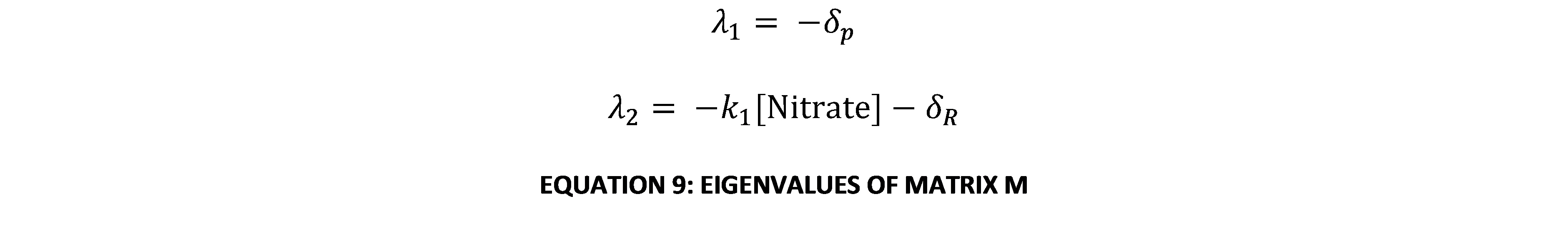Team:BCCS-Bristol/Modelling/GRN/Derivation
From 2010.igem.org
(→GRN Models) |
(→GRN Models) |
||
| Line 18: | Line 18: | ||
[[Image:BCCS_GRN_EQ_7.jpg|frameless|center|upright=4|BCCS-Bristol GRN equation 7]] | [[Image:BCCS_GRN_EQ_7.jpg|frameless|center|upright=4|BCCS-Bristol GRN equation 7]] | ||
| + | |||
| + | This can be written in matrix form to help classify the dynamics of the system. The eigenvalues of the matrix form, M, determine the system’s dynamic states. | ||
| + | |||
| + | [[Image:BCCS_GRN_EQ_8.jpg|frameless|center|upright=4|BCCS-Bristol GRN equation 8]] | ||
| + | |||
| + | The eigenvalues of this matrix are: | ||
| + | |||
| + | [[Image:BCCS_GRN_EQ_9.jpg|frameless|center|upright=4|BCCS-Bristol GRN equation 9]] | ||
| + | |||
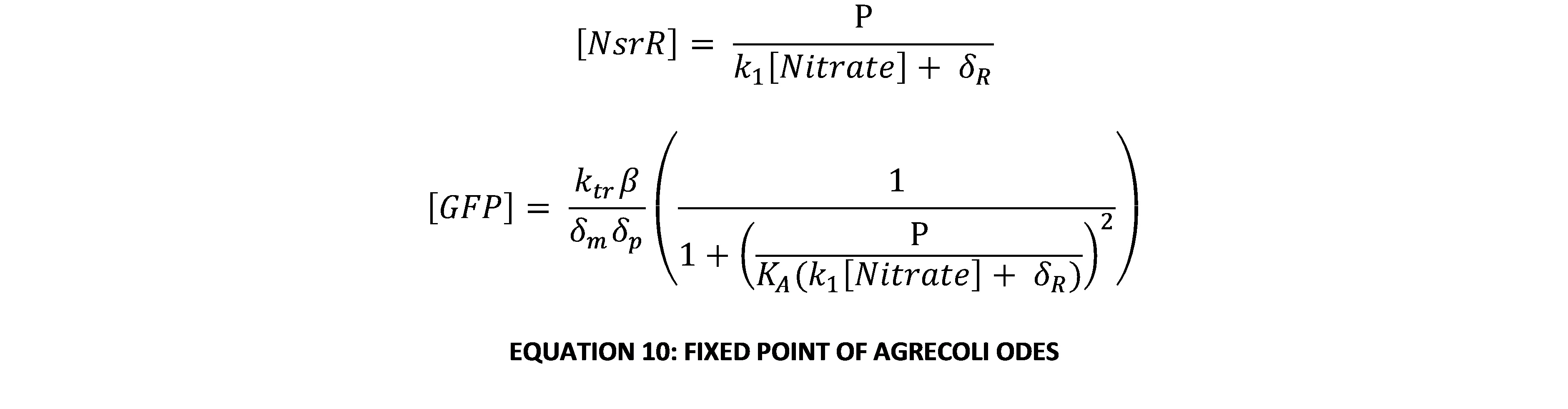
| + | Since all the constants in this system of equations are constrained to be real semi-positive numbers (i.e. ≥0), both eigenvalues are always real and negative. This means that the only fixed point on the phase-plane will be a stable node (possibly degenerate or improper). Further, it is impossible for either eigenvalue to be 0 as both breakdown constants of GFP and NsrR are known to be explicitly positive (i.e. >0). Thus it is clear that the system always converges to a single equilibrium point. By finding the nullclines of the equation and working out where they meet, the fixed point can be found: | ||
| + | |||
| + | [[Image:BCCS_GRN_EQ_10.jpg|frameless|center|upright=4|BCCS-Bristol GRN equation 10]] | ||
Revision as of 15:57, 18 October 2010
iGEM 2010
Derivation
Discussion of how we derived GRN model....
GRN Models
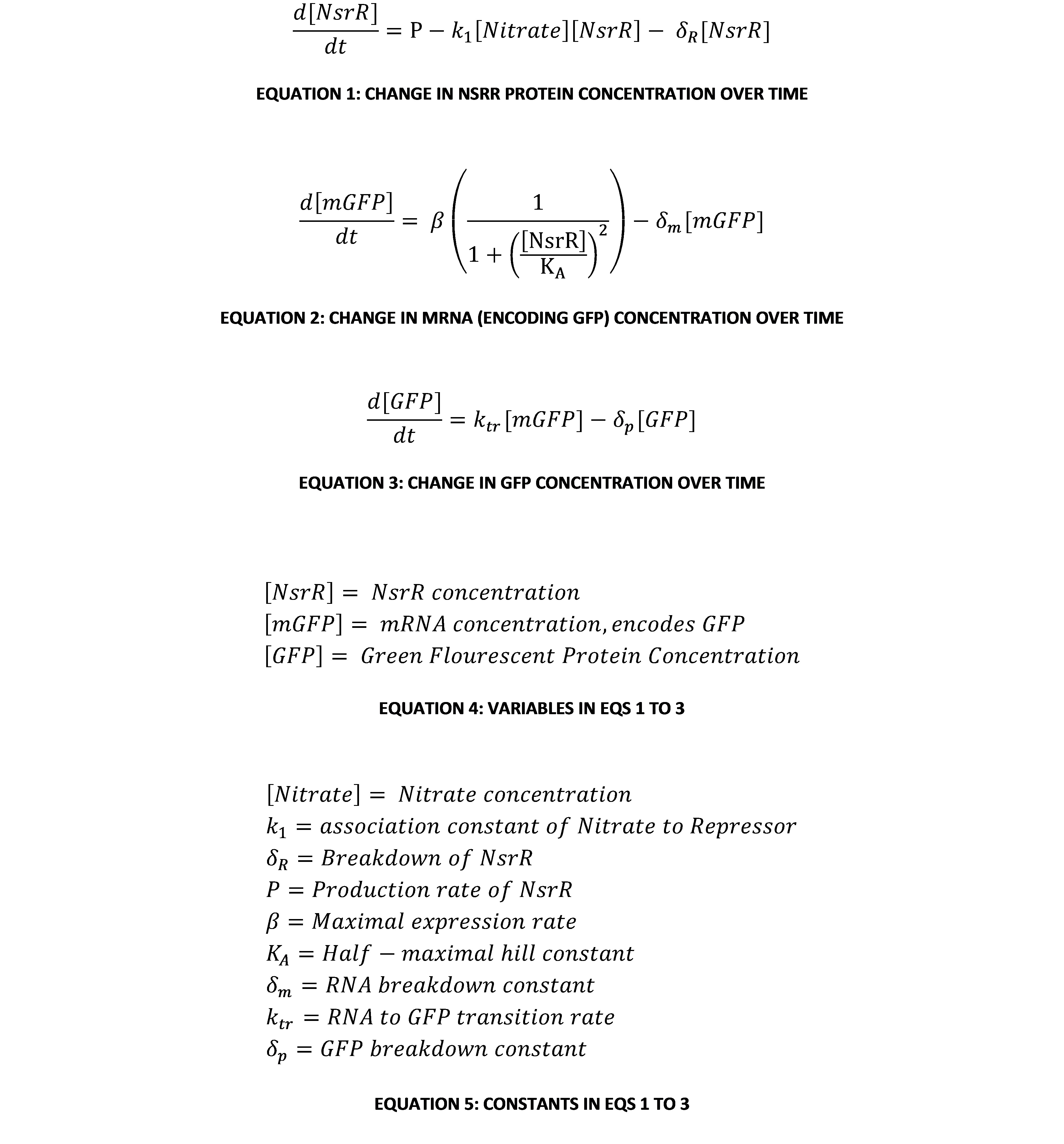
A Gene Regulatory Network (GRN) model is an established way of representing the biochemical reactions within a bacterial cell (7). Representing a network of interacting components using differential equations can be very useful, mainly because one can make analytical and qualitative assessments of a system’s behaviour with very limited information on parameters. This GRN represents a protein called NsrR that represses the production of mRNA encoding GFP, and so indirectly repressing the production of GFP. NsrR can bind to nitrates, effectively destroying its ability to repress the system.
The most interesting relationship in this system of equations is the relationship between the concentration of NsrR repressor protein and the concentration of GFP in the system. A useful way of visualising and analysing the system’s dynamics is using a phase plane portrait in 2 dimensions. However, this means eliminating one of the variables. This can be done by making an equilibrium assumption about the amount of mRNA encoding GFP (mGFP for short). To do this, we set Equation 2 equal to 0. Rearranging in terms of [mGFP], this gives:
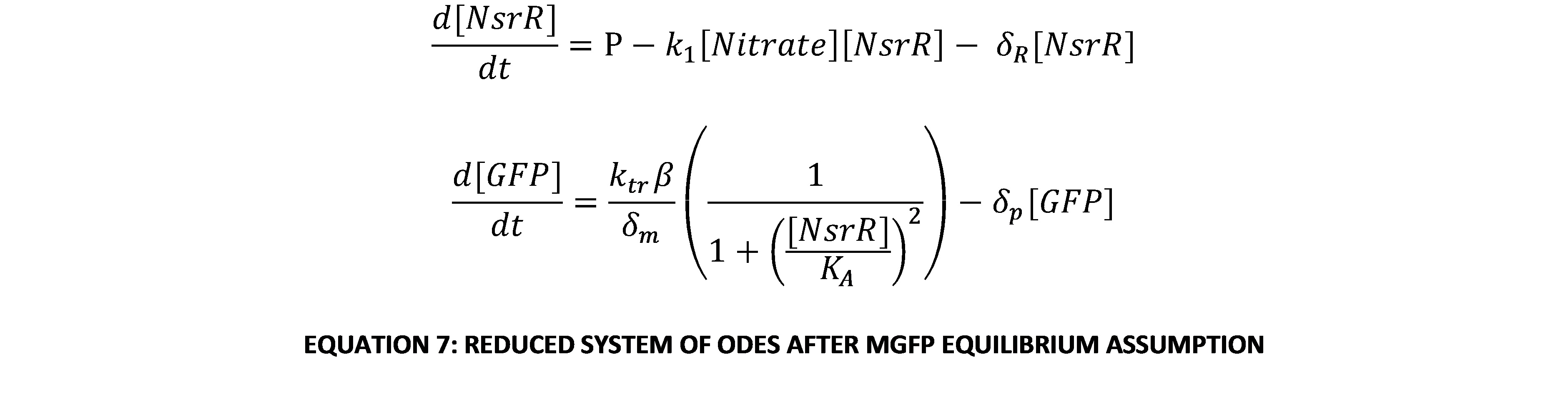
This can be substituted into Equation 3 to give an expression of [GFP] in terms of only itself and [NsrR]. This is now a system of 2 variables, so with a little further manipulation it can be put into a phase-plane portrait. The system of 2 linear ODEs is:
This can be written in matrix form to help classify the dynamics of the system. The eigenvalues of the matrix form, M, determine the system’s dynamic states.
The eigenvalues of this matrix are:
Since all the constants in this system of equations are constrained to be real semi-positive numbers (i.e. ≥0), both eigenvalues are always real and negative. This means that the only fixed point on the phase-plane will be a stable node (possibly degenerate or improper). Further, it is impossible for either eigenvalue to be 0 as both breakdown constants of GFP and NsrR are known to be explicitly positive (i.e. >0). Thus it is clear that the system always converges to a single equilibrium point. By finding the nullclines of the equation and working out where they meet, the fixed point can be found:
 "
"