|
|
| (24 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | {{:Team:BCCS-Bristol/Header}} | | {{:Team:BCCS-Bristol/Header}} |
| | + | __NOTOC__ |
| | + | <html><center> |
| | + | <div style="width:64em;"> |
| | + | <img src="https://static.igem.org/mediawiki/2010/b/bc/AgrEcoli_alt_logo.png" width="100%"/> |
| | + | </div> |
| | + | </center> |
| | + | </html> |
| | | | |
| | + | <center> |
| | + | |
| | + | '''agrEcoli is a device based on modified''' '''''E.coli''''' '''bacteria that detects and signals the presence of nitrates. This allows farmers to map the nutrient content of their fields and optimize their fertiliser use. For more information, see our [https://2010.igem.org/Team:BCCS-Bristol/Project Project Abstract]''' |
| | + | |
| | + | ==Achievements== |
| | + | </center> |
| | + | |
| | + | ===Wetlab=== |
| | | | |
| | {| | | {| |
| - | |style="vertical-align:top;"| | + | | |
| - | <html><style type="text/css">
| + | |
| - | .intro { margin-left:0px; margin-top:10px; padding:10px; border-left:solid 5px #ECF8E0; border-right:solid 5px #ECF8E0; text-align:justify;background:#F5FBEF; }
| + | |
| - | h1 { font-family: 'Ariel Rounded MT Bold', sans-serif;}
| + | |
| - | .intro h1 { font-weight:bold; font-size:medium; border:none; margin:none; padding:none; }
| + | |
| - | .firstHeading { display:none; }
| + | |
| - | #contentSub { display:none; }
| + | |
| - | </style></html>
| + | |
| - | <div class="intro">
| + | |
| - | <h1>Soil Indicator Project</h1>
| + | |
| - | Many crop types need to be harvested and replanted from scratch on an annual basis. To maintain nutrient levels in the soil, farmers often have to put down considerable quantities of fertiliser. This is costly, and a fair proportion of it ends up landing on soil that has enough nutrients anyway. Additionally, some of the nutrients in the fertiliser are often washed away, affecting local ecosystems. This process is called Eutrophication. This is a major environmental concern, as it can cause algal blooms that drain oxygen out of rivers and lakes, killing fish and other wildlife.
| + | |
| | | | |
| - | Our project aims to create a cheap, versatile soil fertility sensor to be used primarily by farmers. Our device will work by sensing whether nutrients in newly ploughed soil are above a given threshold. If this is the case, it will express a colour signal, identifying its area as high in nutrients. Our device will be based on an E-Coli chassis.
| + | * '''A Well Characterised New BioBrick''' |
| | | | |
| - | </div>
| + | :Our new BioBrick ([http://partsregistry.org/Part:BBa_K381001 BBa_K381001]) causes GFP expression in the presence of nitrates. |
| - | |https://static.igem.org/mediawiki/2010/6/6a/Final_Front_page_image.jpg
| + | * '''Elegant Solution to Signal Calibration''' |
| | | | |
| | + | :By using constitutive RFP expression as a baseline, we have found a reliable and accurate way of quantifying nitrate levels in soil. |
| | + | * '''Novel Use of Cell Encapsulation''' |
| | + | |
| | + | :By encapsulating our bacteria in gellan beads, we can keep our bacteria contained and concentrated. This improves visibility on soil, and enhances the environmental safety of our device. |
| | + | * '''Characterising a Pre-Existing BioBrick''' |
| | + | |
| | + | :To better inform our own work, and to add knowledge to the BioBrick Registry, we have characterised Edinburgh 2009’s PyeaR BioBrick ([http://partsregistry.org/Part:BBa_K216009 BBa_ K216009]). |
| | + | |
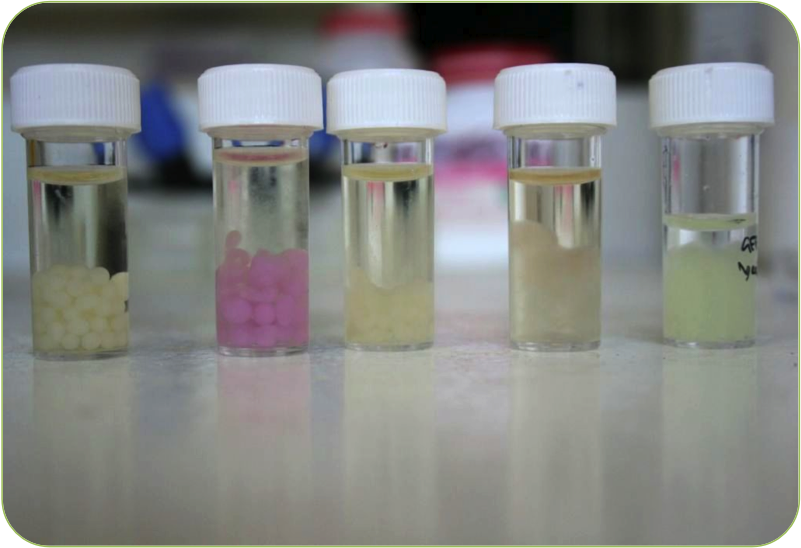
| | + | |[[Image:Beads.png|thumbnail|400px|right|Our new beads]] |
| | |} | | |} |
| | | | |
| | + | {| |
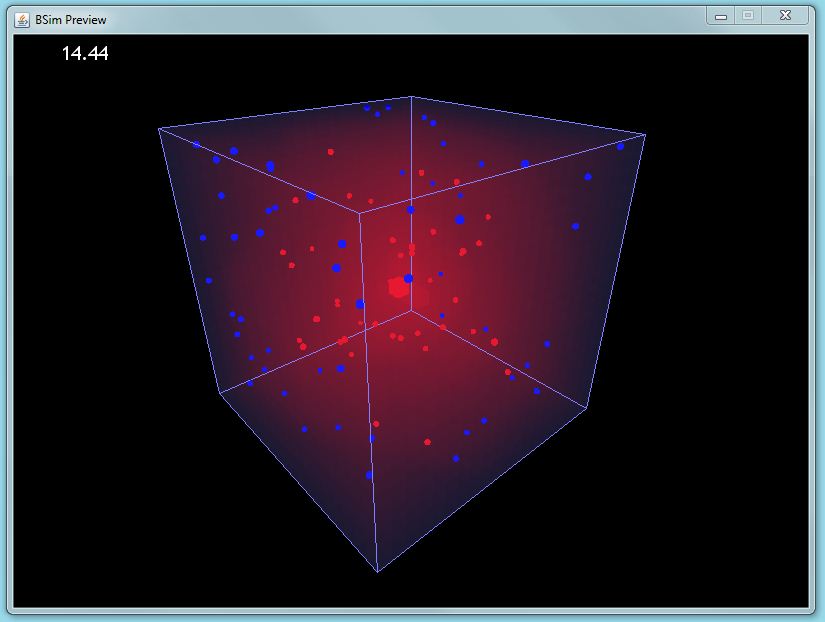
| | + | |[[Image:GUI preview sample.JPG|thumbnail|400px|left|BSim screenshot]] |
| | + | [[Image:Depth0.png|thumbnail|400px|left|Gel strand rendered in BSim]] |
| | | | |
| - | ==Old Project Page (soon to be deleted)==
| |
| | | | |
| | + | | |
| | + | ===Modelling=== |
| | | | |
| - | <html>
| + | *'''BSim Environmental Interactions''' |
| - | <br />
| + | |
| - | <h1>Soil indicator</h1>
| + | :We have extended BSim, our agent-based modelling framework, to model interactions between bacteria and their environment. We have added 3-Dimensional mesh structures to our simulations, and added an adaptive chemical field routine that can solve partial differential equations in an arbitrary 3-D space without any risk of numerical instability. |
| - | <br />
| + | *'''BSim Graphical User Interface (GUI)''' |
| - | <h2>Mission Statement</h2>
| + | |
| - | <p align="justify">Our project aims to create a cheap, versatile soil fertility sensor to be used primarily by farmers. Our device will work by sensing whether nutrients in newly ploughed soil are above a given threshold. If this is the case, it will express a colour signal, identifying its area as high in nutrients. Our device will be based on an E-Coli chassis, as this allows us the greatest flexibility in design. We intend to implement a simple version of this device in the lab, and then characterise it thoroughly as part of the modelling section of this project. The modelling section will also involve more complicated designs, and an investigation of how our bacteria can be grown, deployed, measured and killed in the field.</p>
| + | :We have made BSim more widely accessible by creating a user-friendly and intuitive graphical user interface. This makes BSim accessible to the entire synthetic biology community, rather than just those with JAVA programming knowledge. |
| - | <h2>Motivation</h2>
| + | *'''Gene Regulatory Network Modelling''' |
| - | <p align="justify">Many crop types need to be harvested and replanted from scratch on an annual basis. To maintain nutrient levels in the soil, farmers often have to put down considerable quantities of fertiliser. This is costly, and a fair proportion of it ends up landing on soil that has enough nutrients anyway. Additionally, some of the nutrients in the fertiliser are often washed away, affecting local ecosystems. This process is called eutrophication. This is a major environmental concern, as it can cause algal blooms that drain oxygen out of rivers and lakes, killing fish and other wildlife.
| + | |
| - | Our device is cheaper and more environmentally friendly than chemical nutrient detection systems, and can eventually be used directly on fields. This allows farmers to skip fertilising areas that are already nutrient-rich, saving them money on fertiliser. Less fertiliser on the fields means less nutrient runoff, and consequently less environmental damage. Finally, on a national level, countries can potentially produce more crops with the same quantity of resources. This could be very helpful to poorer nations with growing populations.</p>
| + | :We have investigated the behaviour of our bacteria by creating a mathematical model of their behaviour. This model could then be analysed using numerical and analytic methods. |
| - | <h2>How does it work?</h2>
| + | *'''Collaboration''' |
| - | <p align="justify">Our detector consists of a population of E-coli bacteria which have been modified to detect the presence a set of nutrients. When they sense that the quantity of a nutrient is above a certain threshold, they express a colour (GFP).
| + | |
| - | Many crops, for instance maize or wheat, need to be planted in open soil, grow for an entire season and then be harvested completely out of the ground, leaving an open field for the next planting season. Our plan is to use the detection system immediately before planting a new crop. After a harvest, a farmer would generally plough the field. This is a process by which the topsoil of the field is replaced by the layer of soil immediately beneath it. This is done because topsoil is generally not very good for planting seeds in. This is because it is harder set and lower in nutrients. The newly ploughed soil is representative of the deeper soil in the area, so we intend to use it as the surface on which our detector works.
| + | :Our modelling team have helped to simulate the UCL 2010 team’s new system. Working with another team has also helped to inform the design of the BSim GUI. |
| - | Our bacteria can be grown in quantity and then sprayed across the newly ploughed soil. The colour that the bacteria express may be difficult to see with the naked eye, so we recommend a more mechanised method. An ultraviolet light source can be fitted underneath the tractor or vehicle used to apply fertiliser to the field. This UV light will cause the bacteria to luminesce, and this luminescence can be detected by a camera on the tractor. The fertiliser being deployed by the tractor can then be stopped whenever it detects it is travelling over ground that is already high in nutrients. By this method, the use of fertiliser is limited only to areas with low levels of nutrients.</p> | + | *'''agrEcoli Cost estimates''' |
| - | </html> | + | |
| | + | :In support of our human practices work, our modelling team have looked into the cost to farmers of using agrEcoli, and how much money and fertiliser they can expect to save. |
| | + | |
| | + | |} |
| | + | |
| | + | {| |
| | + | | |
| | + | ===Human Practices=== |
| | + | |
| | + | *'''Publicising agrEcoli''' |
| | + | |
| | + | :Our new approach to human practices, building on previous work by iGEM teams, is a publicity campaign. By presenting our prototype as a functioning and marketable product, we've framed a hypothetical situation in which our project could be released. |
| | + | |
| | + | *'''Public Engagement''' |
| | + | |
| | + | :Our team has visited a school and spoken on two radio shows in order to communicate our ideas with the general public. |
| | + | |
| | + | |[[Image:BCCS-Bristol-ClevedonVisit-03.jpg|thumbnail|400px|right|Public Engagement- Clevedon School Visit]] |
| | + | |} |
| | + | |
| | + | |
| | + | <center> |
| | + | |
| | + | ==Sponsors== |
| | + | [[Image:BCCS_Sponsors_Logo_Banner.png|upright= 3.3|frameless|center]] |
| | + | </center> |

 "
"