Team:Alberta/biobyte2
From 2010.igem.org
(→All BioBytes are stored in TE buffer.) |
|||
| (8 intermediate revisions not shown) | |||
| Line 12: | Line 12: | ||
<div id="horiz-line"></div> | <div id="horiz-line"></div> | ||
| - | BioBytes 2.0 is the heart of the GENOMIKON kit. BioBytes 2.0 is a method of creating novel plasmids through the sequential addition of functional units of DNA. It is designed to be used in a high school setting but has potential | + | BioBytes 2.0 is the heart of the GENOMIKON kit. BioBytes 2.0 is a method of creating novel plasmids through the sequential addition of functional units of DNA. It is designed to be used in a high school setting but has potential applications in professional settings as well. The assembly method we have created has some conceptual similarities to the original BioBytes Assembly System of the 2009 Alberta iGEM project. However, there are some striking differences between the two systems. |
==Traditional Methods== | ==Traditional Methods== | ||
<div id="horiz-line"></div> | <div id="horiz-line"></div> | ||
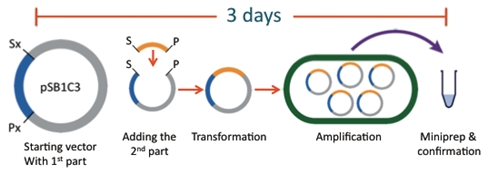
| - | The current assembly standard is the BioBrick method. While the | + | The current assembly standard is the BioBrick method. While the Registry of Parts and the assembly standard have allowed for effective construction of plasmids in a laboratory setting, it has numerous limitations prohibiting its use in high schools. For example, common laboratory protocols such as transformations, ligations, and restriction digests require expensive materials and equipment not available to high schools. Another disadvantage is that it takes from days to weeks to assemble a complicated construct using this method. An experiment of such length far surpasses the average high school student’s attention span and time allotted in a curriculum. |
| - | [[Image:team-alberta-biobrick-tour.jpg|center|frame|Figure 1.1. Traditional BioBrick construction | + | [[Image:team-alberta-biobrick-tour.jpg|center|frame|Figure 1.1. Traditional BioBrick construction takes approximately 3 days to complete the addition of one BioBrick; from the digestion of the original constructs to transformation and confirmation.]] |
====Comparison==== | ====Comparison==== | ||
| - | The BioBytes | + | The BioBytes 2.0 assembly method has provided a solution to these issues. It is fast. The addition of one BioByte to a construct takes under 10 minutes. Therefore, creating a plasmid of desired specifications can happen in an afternoon, rather than the 3 or more days to create a plasmid through the traditional BioBrick method. It is accessible. The GENOMIKON kit is completely self-contained, requiring no additional equipment or reagents other than a hot plate and a beaker. This eliminates the need for the expensive equipment and reagents found in a traditional university laboratory. |
| - | The addition of one | + | |
[[Image:team-alberta-building-tour.jpg|center|frame|BioByte version 2.0 construction.]] | [[Image:team-alberta-building-tour.jpg|center|frame|BioByte version 2.0 construction.]] | ||
| Line 30: | Line 30: | ||
<div id="horiz-line"></div> | <div id="horiz-line"></div> | ||
| - | + | BioBytes 2.0 is composed of three main components. The construct begins with an anchor-Byte piece which is attached to an iron micro bead. Because of the magnetic nature of these beads, they can be positioned by using a simple magnet. The BioBytes are added to the anchor-Byte one-at-a-time, in sequence. This is possible due to the alternating overhang structure of the BioBytes. Finally, a cap Byte is added allowing for circularization of the construct. No ligation is required for circularization. The construct is now ready to transform. | |
| + | ====Iron Micro Beads==== | ||
| + | Iron micro beads purchased from New England Biolabs have covalently attached poly-T tails. The bead allows us to orient the DNA with magnets, making washing and subsequent attachments easier. | ||
| - | ====Anchor Byte==== | + | ====Anchor-Byte==== |
| - | The | + | The anchor is a part that attaches directly onto the iron micro beads. It is composed of: |
*a poly-A tail | *a poly-A tail | ||
| Line 44: | Line 46: | ||
[[Image:team-alberta-anchor-tour.jpg|center|frame|The two components of the anchoring system.]] | [[Image:team-alberta-anchor-tour.jpg|center|frame|The two components of the anchoring system.]] | ||
| - | + | We have designed several anchor-Byte constructs which can begin the process of assembly. An anchor-Byte is comprised of an anchor piece ligated to the first Byte of the assembly, usually a selectable marker. This allows for complete constructs to be selected for. As well, the incorporation of a BsaI cut site into the anchor, before the first Byte, gives versatility to the construct because the first Byte and the rest of the construct can be removed from the anchor, and used as a Byte on its own. This allows for multiplexing experiments. | |
| - | + | Once the selectable marker is ligated to the first Byte, we anneal the anchor-Byte to the poly-T tails on the iron micro beads. We have created anchors with both AB and BA ends so that assemblies can begin with any type of Byte. | |
| + | The results of one of our experiments is shown here. Note that the interaction between the anchor and the bead is non-covalent. The anchor along with the construct can be separated from the bead with heat. The gel shows the process of anchoring a construct. An anchor-Byte construct of 1kb was allowed to anneal to the magnetic beads. This was done in excess, and the supernatant is shown in the first lane. A subsequent wash step showed the absence of DNA, indicating that DNA construct is stably bound. The construct was melted from the scaffold at 75 degrees Celsius. The melted construct can be seen in the last lane. | ||
| - | + | [[Image:Alberta_AnchorPiece.png|500px|center|frame|Figure of the anchor system with the BioBytes 2.0 Assembly System.]] | |
| - | + | ||
| - | |||
| - | |||
| - | |||
====BioBytes 2.0==== | ====BioBytes 2.0==== | ||
| - | + | The BioBytes 2.0 assembly method has a number of unique features. Each Byte has 4-base 5' overhangs on each side. These overhangs have been engineered to serve a number of purposes. The overhangs allow for ordered and sequential addition of parts to growing constructs. | |
[[Image:team-alberta-bytes-tour.jpg|500px|center|thumb|BioBytes come in two flavors: 'AB' and 'BA'.]] | [[Image:team-alberta-bytes-tour.jpg|500px|center|thumb|BioBytes come in two flavors: 'AB' and 'BA'.]] | ||
| - | The addition of each | + | The addition of each Byte to a growing construct takes about 7 minutes. |
====Cap==== | ====Cap==== | ||
| - | The | + | The cap Byte is analogous to the anchor. The cap finishes the construct, where the anchor starts a construct. The cap is comprised of a poly-T tail, complimentary to the poly-A tail of the anchor piece. The cap also contains a BsaI cut site which allows its removal from the construct. It can follow an AB or BA format. |
[[Image:team-alberta-cap-tour.jpg|center|The cap. The poly-T anneals to the poly-A of the anchor component to circularize the complete plasmid.]] | [[Image:team-alberta-cap-tour.jpg|center|The cap. The poly-T anneals to the poly-A of the anchor component to circularize the complete plasmid.]] | ||
| - | When finishing a construct, the cap is | + | When finishing a construct, the cap is ligated to the growing construct, in the same manner as another Byte. The completed construct is then heated, to melt the anchor piece from the iron micro bead. The solution is removed from the beads and allowed to cool. The cap and anchor then have opportunity to anneal to each other. The construct is now ready for transformation without ligation. |
| - | == | + | ==BioBytes 2.0 Assembly Process== |
<div id="horiz-line"></div> | <div id="horiz-line"></div> | ||
[[Image:team-alberta-biobyteprocess-tour.jpg|center|frame|The entire construction process can be completed in an afternoon.]] | [[Image:team-alberta-biobyteprocess-tour.jpg|center|frame|The entire construction process can be completed in an afternoon.]] | ||
| - | |||
| - | |||
===Please note that the GENOMIKON kit comes with premade anchor-Byte constructs.=== | ===Please note that the GENOMIKON kit comes with premade anchor-Byte constructs.=== | ||
| - | === | + | ===The GENOMIKON kit supplies plastic micropipettes than can dispense 25ul per drop.=== |
| + | |||
:# Mix the iron micro beads for 10 minutes. | :# Mix the iron micro beads for 10 minutes. | ||
| - | :# Transfer a 25 ul (one drop) aliquot of iron micro beads to a 1.5 mL tube | + | :# Transfer a 25 ul (one drop) aliquot of iron micro beads to a 1.5 mL tube. |
:# Pull the beads to the side using the magnetic tube rack supplied. | :# Pull the beads to the side using the magnetic tube rack supplied. | ||
:# Remove and discard the supernatant. | :# Remove and discard the supernatant. | ||
| Line 92: | Line 90: | ||
:# Repeat steps 5 to 7. | :# Repeat steps 5 to 7. | ||
:# Add 200 ng of the next Byte of your construct, making sure that a BA Byte follows an AB Byte and vice versa. Add an appropriate amount of 2x QuickLigase Buffer, Quick Ligase and TE to a total volume of 25ul. | :# Add 200 ng of the next Byte of your construct, making sure that a BA Byte follows an AB Byte and vice versa. Add an appropriate amount of 2x QuickLigase Buffer, Quick Ligase and TE to a total volume of 25ul. | ||
| - | Example: | + | ::Example: |
{|style="margin-left:40px;" | {|style="margin-left:40px;" | ||
! Reagent !! Volume (ul) | ! Reagent !! Volume (ul) | ||
| Line 108: | Line 106: | ||
| - | : | + | ::14. Flick gently to resuspend. Allow ligation for five minutes, flicking gently every minute. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ::15. Add 25 ul of Wash Buffer to the tube. Flick gently. | ||
| - | Using this | + | ::16. Repeats steps 6 and 7. |
| + | |||
| + | ::17. Repeat steps 5 to 7 twice. | ||
| + | |||
| + | ::18. Repeat steps 13 to 17 for each subsequent Byte addition, including the last Byte. If the last Byte is cap, it must be added in 20 X molar excess. | ||
| + | |||
| + | ::19. After the last Byte has been ligated and washed, addition of 25 ul of 75C Elution Buffer. The entire tube should be kept at 75C for ten minutes to allow the DNA construct to elute off the beads. | ||
| + | |||
| + | ::20. After ten minutes, put the tube into a magnetic rack in the 75C waterbath. Allow the beads to be pulled aside and collect the supernatant into a clean 1.5 mL tube. The supernatant will contain your construct. | ||
| + | |||
| + | |||
| + | Using this protocol, we were able to assemble an octamer in an afternoon! | ||
==Byte Design== | ==Byte Design== | ||
<div id="horiz-line"></div> | <div id="horiz-line"></div> | ||
| - | Because the overhangs in this year's project are only 4 bases long, and because the ends are malleable, | + | Because the overhangs in this year's project are only 4 bases long, and because the ends are malleable, they give additional versatility. In this classification system, AB parts are "ORF" parts. The "ORF" parts contain the coding sequence of genes. However, these "ORF" parts do not contain a complete open reading frame. They lack a promoter, a RBS, a start codon, a stop codon and a terminator. Therefore, an "ORF" part by itself has no function at all. We have coined this type of parts "naked ORFs" |
| - | The BA parts that can be ligated to the "ORF" parts will reconstitute these functions. For this reason BA parts are now termed "Linker" Parts. The "Linkers" can reconstitute the start and stop codons, the promoters and terminators, but they can also introduce, protein fusion linkers and protein tags. This give this year's | + | The BA parts that can be ligated to the "ORF" parts will reconstitute these functions. For this reason BA parts are now termed "Linker" Parts. The "Linkers" can reconstitute the start and stop codons, the promoters and terminators, but they can also introduce, protein fusion linkers and protein tags. This give this year's BioBytes 2.0 design added flexibility and potential for testing regulatory regions of DNA. |
| - | + | [[Image:Alberta_BioBytesProteinLinker.png|250px|center|frame|The BioByte 2.0 ends are designed to be adaptable to protein linkers.]] | |
| Line 132: | Line 136: | ||
<div id="horiz-line"></div> | <div id="horiz-line"></div> | ||
| - | New Biobytes are constructed with the aid of our Base Plasmids v.2. The Base Plasmids v.2 have an RFP coding cassette (J04450) flanked by BsaI cut sites that leave the appropriate overhangs to create either | + | New Biobytes are constructed with the aid of our Base Plasmids v.2. The Base Plasmids v.2 have an RFP coding cassette (J04450) flanked by BsaI cut sites that leave the appropriate overhangs to create either an AB part or a BA part. Because the BsaI restriction enzyme cuts at a site downstream of the recognition sequence, the overhang sequence can be engineered to be unique. We have designed our own overhangs that define the AB and the BA parts. |
| + | |||
| + | |||
| + | To create a new part PCR the part of interest, incorporating BsaI cut sites with appropriate overhangs onto each side. Then, cut the Base Plasmid v.2 and the PCR product with BsaI. Ligate it together. Again, because the overhangs are unique, the plasmid backbone can not re-ligate without an insert. The insert can only be the new part of interest, or the RFP coding cassette that was originally in the base plasmid, leading to a total of 2 possible ligation products. When transformed, the ligation products can appear red (if the original RFP cassette is reinserted) or white (if the part of interest is inserted). This provides a selection method for plasmids that have the part of interest in it. | ||
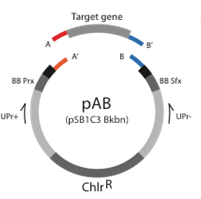
[[Image:Alberta_pAB_Plasmid.png|350px|center|frame|Figure Base Plasmid v.2. The Base plasmids can be cut leaving unique overhangs so that directional cloning of new parts can be inserted. The lack of the RFP coding cassette provides a selection method for colonies containing new parts]] | [[Image:Alberta_pAB_Plasmid.png|350px|center|frame|Figure Base Plasmid v.2. The Base plasmids can be cut leaving unique overhangs so that directional cloning of new parts can be inserted. The lack of the RFP coding cassette provides a selection method for colonies containing new parts]] | ||
| - | Because all the parts are created in the same plasmid, mass production of parts for the kit is also a straight forward matter. Using universal primers of our own design, which start about 100 base before the BsaI cut sites. The PCR is then digested with BsaI. Starting the PCR 100 bases outside the part allows for us to check that the part is completely digested. Before parts are used in assembly, they are purified using weak anion exchange column through HPLC (high performance liquid chromatography). | + | Because all the parts are created in the same plasmid, mass production of parts for the kit is also a straight forward matter. Using universal primers of our own design, which start about 100 base before the BsaI cut sites. The PCR is then digested with BsaI. Starting the PCR 100 bases outside the part allows for us to check that the part is completely digested. Before parts are used in assembly, they are purified using a weak anion exchange column through HPLC (high performance liquid chromatography). |
[[Image:Alberta_PCR.png|350px|center|frame|Figure Part Mass Production. New parts are PCRed, Digested with BsaI, then purified through an HPLC before being used in assembly]] | [[Image:Alberta_PCR.png|350px|center|frame|Figure Part Mass Production. New parts are PCRed, Digested with BsaI, then purified through an HPLC before being used in assembly]] | ||
| Line 143: | Line 150: | ||
<div id="horiz-line"></div> | <div id="horiz-line"></div> | ||
| - | + | There are a few similarities between the original BioBytes 1.0 assembly method of Team Alberta 2009 and the new BioBytes 2.0 system that we have put forward this year. However, if you look closely, there are a number of profound differences between the systems. | |
| + | |||
| + | The similarities in the systems reside in the theory of alternating and complimentary ends. The AB and BA parts can only ligate to each other, and AB parts can not ligate to themselves and BA parts can not ligate to themselves. However, this year we are working with 4-basepair overhangs, not the 12-basepair overhangs used last year. Also, this year we are ligating they bytes together, whereas last year they were not ligated. | ||
| - | The | + | The iron micro beads with the poly-T tail are also unique to this year. Last year, the constructs were anchored using a biotin and streptavidin process requiring an enzyme to cleave the construct. This means that the release and circularizing of the constructs of each year are unique processes. |
| - | The | + | [[Image:Alberta_BioByte2.0vs1.0.png|250px|center|frame|The similarities and differences between the BioByte 2.0 and last year's BioByte 1.0]] |
The "BioBytes Version 2.0" construction method has been shown to create plasmids from up to 8 separate parts in | The "BioBytes Version 2.0" construction method has been shown to create plasmids from up to 8 separate parts in | ||
Latest revision as of 03:21, 28 October 2010
Overview
BioBytes 2.0 is the heart of the GENOMIKON kit. BioBytes 2.0 is a method of creating novel plasmids through the sequential addition of functional units of DNA. It is designed to be used in a high school setting but has potential applications in professional settings as well. The assembly method we have created has some conceptual similarities to the original BioBytes Assembly System of the 2009 Alberta iGEM project. However, there are some striking differences between the two systems.
Traditional Methods
The current assembly standard is the BioBrick method. While the Registry of Parts and the assembly standard have allowed for effective construction of plasmids in a laboratory setting, it has numerous limitations prohibiting its use in high schools. For example, common laboratory protocols such as transformations, ligations, and restriction digests require expensive materials and equipment not available to high schools. Another disadvantage is that it takes from days to weeks to assemble a complicated construct using this method. An experiment of such length far surpasses the average high school student’s attention span and time allotted in a curriculum.
Comparison
The BioBytes 2.0 assembly method has provided a solution to these issues. It is fast. The addition of one BioByte to a construct takes under 10 minutes. Therefore, creating a plasmid of desired specifications can happen in an afternoon, rather than the 3 or more days to create a plasmid through the traditional BioBrick method. It is accessible. The GENOMIKON kit is completely self-contained, requiring no additional equipment or reagents other than a hot plate and a beaker. This eliminates the need for the expensive equipment and reagents found in a traditional university laboratory.
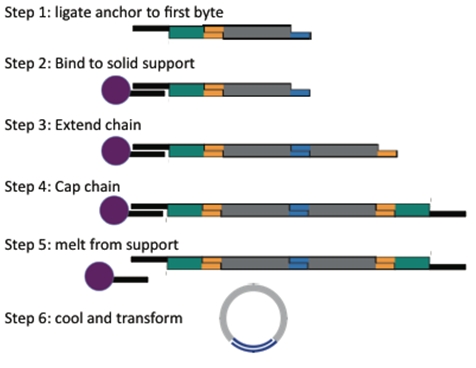
Components of the System
BioBytes 2.0 is composed of three main components. The construct begins with an anchor-Byte piece which is attached to an iron micro bead. Because of the magnetic nature of these beads, they can be positioned by using a simple magnet. The BioBytes are added to the anchor-Byte one-at-a-time, in sequence. This is possible due to the alternating overhang structure of the BioBytes. Finally, a cap Byte is added allowing for circularization of the construct. No ligation is required for circularization. The construct is now ready to transform.
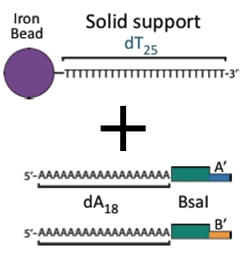
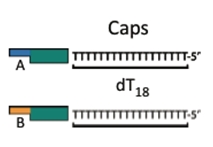
Iron Micro Beads
Iron micro beads purchased from New England Biolabs have covalently attached poly-T tails. The bead allows us to orient the DNA with magnets, making washing and subsequent attachments easier.
Anchor-Byte
The anchor is a part that attaches directly onto the iron micro beads. It is composed of:
- a poly-A tail
- a BsaI recognition site
- an A or B overhang
We have designed several anchor-Byte constructs which can begin the process of assembly. An anchor-Byte is comprised of an anchor piece ligated to the first Byte of the assembly, usually a selectable marker. This allows for complete constructs to be selected for. As well, the incorporation of a BsaI cut site into the anchor, before the first Byte, gives versatility to the construct because the first Byte and the rest of the construct can be removed from the anchor, and used as a Byte on its own. This allows for multiplexing experiments.
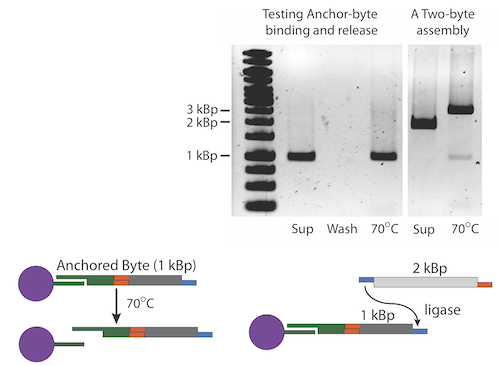
Once the selectable marker is ligated to the first Byte, we anneal the anchor-Byte to the poly-T tails on the iron micro beads. We have created anchors with both AB and BA ends so that assemblies can begin with any type of Byte. The results of one of our experiments is shown here. Note that the interaction between the anchor and the bead is non-covalent. The anchor along with the construct can be separated from the bead with heat. The gel shows the process of anchoring a construct. An anchor-Byte construct of 1kb was allowed to anneal to the magnetic beads. This was done in excess, and the supernatant is shown in the first lane. A subsequent wash step showed the absence of DNA, indicating that DNA construct is stably bound. The construct was melted from the scaffold at 75 degrees Celsius. The melted construct can be seen in the last lane.
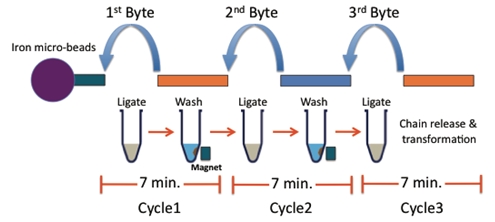
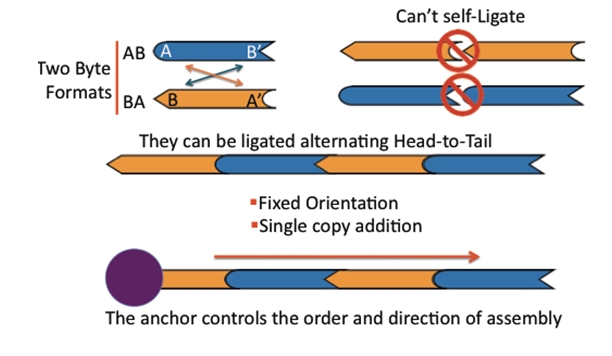
BioBytes 2.0
The BioBytes 2.0 assembly method has a number of unique features. Each Byte has 4-base 5' overhangs on each side. These overhangs have been engineered to serve a number of purposes. The overhangs allow for ordered and sequential addition of parts to growing constructs.
The addition of each Byte to a growing construct takes about 7 minutes.
Cap
The cap Byte is analogous to the anchor. The cap finishes the construct, where the anchor starts a construct. The cap is comprised of a poly-T tail, complimentary to the poly-A tail of the anchor piece. The cap also contains a BsaI cut site which allows its removal from the construct. It can follow an AB or BA format.
When finishing a construct, the cap is ligated to the growing construct, in the same manner as another Byte. The completed construct is then heated, to melt the anchor piece from the iron micro bead. The solution is removed from the beads and allowed to cool. The cap and anchor then have opportunity to anneal to each other. The construct is now ready for transformation without ligation.
BioBytes 2.0 Assembly Process
Please note that the GENOMIKON kit comes with premade anchor-Byte constructs.
The GENOMIKON kit supplies plastic micropipettes than can dispense 25ul per drop.
- Mix the iron micro beads for 10 minutes.
- Transfer a 25 ul (one drop) aliquot of iron micro beads to a 1.5 mL tube.
- Pull the beads to the side using the magnetic tube rack supplied.
- Remove and discard the supernatant.
- Add 50 ul (2 drops) of supplied Wash Buffer to the beads. Flick gently to resuspend.
- Pull the beads to the side using the magnetic tube rack.
- Remove and discard the supernatant.
- Repeat steps 5 to 7.
- Add 200 ng of a premade anchor-Byte construct to the beads and top off the volume to 25ul with the TE Buffer supplied. Flick gently to resuspend.
- Allow annealing for 30 minutes, mixing by flicking every 5 minutes. Ensure that there are no droplets on the sides of the tube.
- Repeat steps 6 and 7.
- Repeat steps 5 to 7.
- Add 200 ng of the next Byte of your construct, making sure that a BA Byte follows an AB Byte and vice versa. Add an appropriate amount of 2x QuickLigase Buffer, Quick Ligase and TE to a total volume of 25ul.
- Example:
| Reagent | Volume (ul) |
|---|---|
| (40ng/ul) AB kan Byte | 5 |
| 2X Quick Ligase Buffer | 13 |
| TE Buffer | 6 |
| Quick Ligase | 1 |
| TOTAL | 25 |
- 14. Flick gently to resuspend. Allow ligation for five minutes, flicking gently every minute.
- 15. Add 25 ul of Wash Buffer to the tube. Flick gently.
- 16. Repeats steps 6 and 7.
- 17. Repeat steps 5 to 7 twice.
- 18. Repeat steps 13 to 17 for each subsequent Byte addition, including the last Byte. If the last Byte is cap, it must be added in 20 X molar excess.
- 19. After the last Byte has been ligated and washed, addition of 25 ul of 75C Elution Buffer. The entire tube should be kept at 75C for ten minutes to allow the DNA construct to elute off the beads.
- 20. After ten minutes, put the tube into a magnetic rack in the 75C waterbath. Allow the beads to be pulled aside and collect the supernatant into a clean 1.5 mL tube. The supernatant will contain your construct.
Using this protocol, we were able to assemble an octamer in an afternoon!
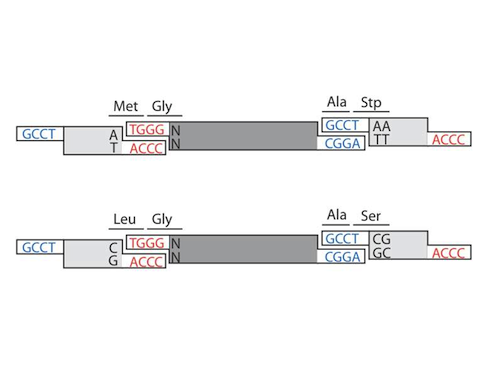
Byte Design
Because the overhangs in this year's project are only 4 bases long, and because the ends are malleable, they give additional versatility. In this classification system, AB parts are "ORF" parts. The "ORF" parts contain the coding sequence of genes. However, these "ORF" parts do not contain a complete open reading frame. They lack a promoter, a RBS, a start codon, a stop codon and a terminator. Therefore, an "ORF" part by itself has no function at all. We have coined this type of parts "naked ORFs" The BA parts that can be ligated to the "ORF" parts will reconstitute these functions. For this reason BA parts are now termed "Linker" Parts. The "Linkers" can reconstitute the start and stop codons, the promoters and terminators, but they can also introduce, protein fusion linkers and protein tags. This give this year's BioBytes 2.0 design added flexibility and potential for testing regulatory regions of DNA.
Byte Construction
New Biobytes are constructed with the aid of our Base Plasmids v.2. The Base Plasmids v.2 have an RFP coding cassette (J04450) flanked by BsaI cut sites that leave the appropriate overhangs to create either an AB part or a BA part. Because the BsaI restriction enzyme cuts at a site downstream of the recognition sequence, the overhang sequence can be engineered to be unique. We have designed our own overhangs that define the AB and the BA parts.
To create a new part PCR the part of interest, incorporating BsaI cut sites with appropriate overhangs onto each side. Then, cut the Base Plasmid v.2 and the PCR product with BsaI. Ligate it together. Again, because the overhangs are unique, the plasmid backbone can not re-ligate without an insert. The insert can only be the new part of interest, or the RFP coding cassette that was originally in the base plasmid, leading to a total of 2 possible ligation products. When transformed, the ligation products can appear red (if the original RFP cassette is reinserted) or white (if the part of interest is inserted). This provides a selection method for plasmids that have the part of interest in it.
Because all the parts are created in the same plasmid, mass production of parts for the kit is also a straight forward matter. Using universal primers of our own design, which start about 100 base before the BsaI cut sites. The PCR is then digested with BsaI. Starting the PCR 100 bases outside the part allows for us to check that the part is completely digested. Before parts are used in assembly, they are purified using a weak anion exchange column through HPLC (high performance liquid chromatography).
BioBytes 2.0 vs. BioBytes
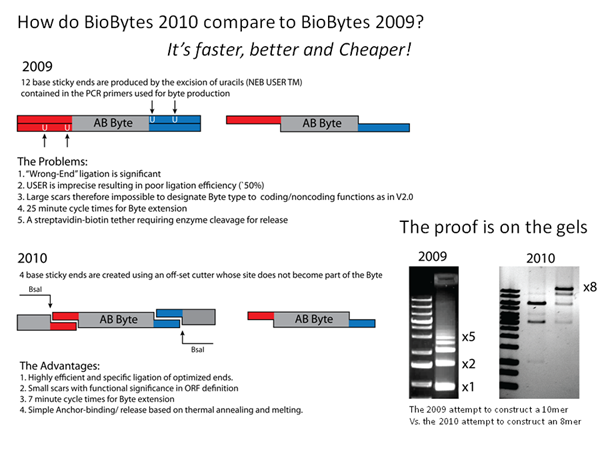
There are a few similarities between the original BioBytes 1.0 assembly method of Team Alberta 2009 and the new BioBytes 2.0 system that we have put forward this year. However, if you look closely, there are a number of profound differences between the systems.
The similarities in the systems reside in the theory of alternating and complimentary ends. The AB and BA parts can only ligate to each other, and AB parts can not ligate to themselves and BA parts can not ligate to themselves. However, this year we are working with 4-basepair overhangs, not the 12-basepair overhangs used last year. Also, this year we are ligating they bytes together, whereas last year they were not ligated.
The iron micro beads with the poly-T tail are also unique to this year. Last year, the constructs were anchored using a biotin and streptavidin process requiring an enzyme to cleave the construct. This means that the release and circularizing of the constructs of each year are unique processes.
The "BioBytes Version 2.0" construction method has been shown to create plasmids from up to 8 separate parts in an afternoon's work. This is a vast improvement.
 "
"