Team:Imperial College London/Modules/Signaling
From 2010.igem.org
| Modules | Overview | Detection | Signaling | Fast Response |
| Our design consists of three modules; Detection, Signaling and a Fast Response, each of which can be exchanged with other systems. We used a combination of modelling and human practices to define our specifications. Take a look at the overview page to get a feel for the outline, then head to the full module pages to find out how we did it. | |
| Signalling Module |
|
The signalling module allows transduction of the signal from the detection module into a response produced by the output module.
Having decided that we wanted to detect the parasite protease, we realised that a possible method might be to have it cleave a protein, which would then result in a downstream signalling pathway. A well-known signalling system using peptides is that of quorum sensing in gram positive bacteria. The signals, or quorum sensing molecules, are known as autoinducing peptides (AIPs) and are transported out of a cell on activation of certain pathways.
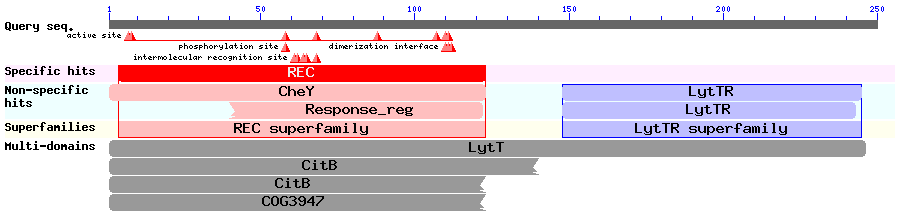
AIPs can remain as linear peptides, or they can be post-translationally modified to become circular. We needed to find a system which signals via a linear AIP, and one such example is the ComCDE system in Streptococcus pneumoniae which signals via a two component signal transduction system. ComC codes for the competence-stimulating peptide-1 (CSP-1) which has the amino acid sequence NH2-EMRLSKFFRDFILQRKK-COOH. CSP-1 is transported out of cells and subsequently detected by the sensory histidine kinase ComD, either belonging to the cell from which it came, or a neighbouring one. ComD then autophosphorylates and then activates the response regulator (RR) ComE by phosphorylating it. ComE then binds to two 9bp imperfect direct repeats to induce transcription of specific target genes. Results of a BLAST search using ComE (Accession no. AAC44897): We ran a BLAST search to ensure there were no homologous systems inherent in B. subtilis that might result in either crosstalk, noise or false positives. The BLAST search also showed that ComE is part of the LytTR superfamily of RRs, and we could therefore try to find out if any of the other members of the superfamily were well characterised, which would give us more information about the mechanism by which ComE works. In 2006, [http://jb.asm.org/cgi/content/full/188/12/4169 Galperin] discussed the structures of various response regulators, including the LytTR superfamily. The LytTR superfamily of RRs binds to DNA via a 'novel' DBD, as is described by [http://nar.oxfordjournals.org/cgi/content/full/30/11/2453?ijkey=ee5882350a787bd60a99afc5778cb7b247a1ea59 Nikolskaya & Galperin]. In 2008, [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2430735/ Sidote et al] describe how they solved the crystal structure of the RR AgrA from S. aureus (a member of the LytTR superfamily) and show how it binds to DNA. Two AgrA proteins bind to DNA at a distance of 12bp from each other, but it is unclear when exactly the dimerisation occurs. The fact that ComE might dimerise before binding to DNA to induce transcription of target genes was important to take into consideration when modelling this system.
In order for us to get a strong enough readout, it was really important to establish the concentration of CSP-1 needed to activate the system. From reading '[http://ukpmc.ac.uk/backend/ptpmcrender.cgi?accid=PMC40587&blobtype=pdf An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae]' by Havarstein, Coomaraswamy & Morrison, we found out that in previous studies only 10ng/ul of CSP-1 was needed to activate the two component signal transduction system. They also comment on the stability of CSP-1 and outlined other parameters affecting response to CSP-1, such as pH and the growth phase of the bacteria. This information was used by the modelling team to calculate how strong our response would be, and whether there was any possibility of getting a false positive.
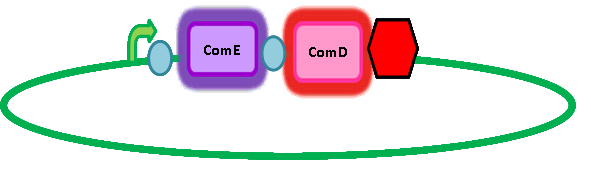
In order to get transcription of TEV protease upon activation of ComE, we had to add a ComE binding site upstream of the TEV protease gene. We read 'Two Separate Quorum-Sensing Systems Upregulate Transcription of the Same ABC Transporter in Streptococcus pneumoniae' by [http://jb.asm.org/cgi/content/full/186/10/3078 Knutsen, Ween & Håvarstein] to determine the ComE binding site. They also assume that ComE binds DNA as a homodimer. The ComE binding sites are described in detail by [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00584.x/full Halfmann, Hakenbeck & Brückner]. They took the ComA and ComC promoters, which each contained a ComE binding site, out of context and used it to initiate transcription of various target genes. However, they integrated this system into S. pneumoniae so we cannot be sure that it will work in B. subtilis. Similarly, there is a possibility that there is a similar transcriptional regulator inherent in B. subtilis which might result in false positives. However, having done homology searches in BLAST, there seems to be no homologs and so we have assumed that there is no risk of false positives arising from a homologous response regulator in B. subtilis. Here's a picture of the final construct: |
 "
"