Team:Imperial College London/Modelling
From 2010.igem.org
(Difference between revisions)
(Changing nomenclature of the amplifier systems. still need to change ilustrations) |
(CHanging diagrams) |
||
| Line 72: | Line 72: | ||
</ol> | </ol> | ||
| - | + | <div ALIGN=CENTER> | |
| - | + | {| style="background:#e7e7e7;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5" width=704px; | |
| - | + | ||
| - | + | ||
| - | {| style=" | + | |
|- | |- | ||
| - | | | + | |[[Image:Wiki_comparison_of_models.png|700px]] |
| - | + | ||
| - | + | ||
|- | |- | ||
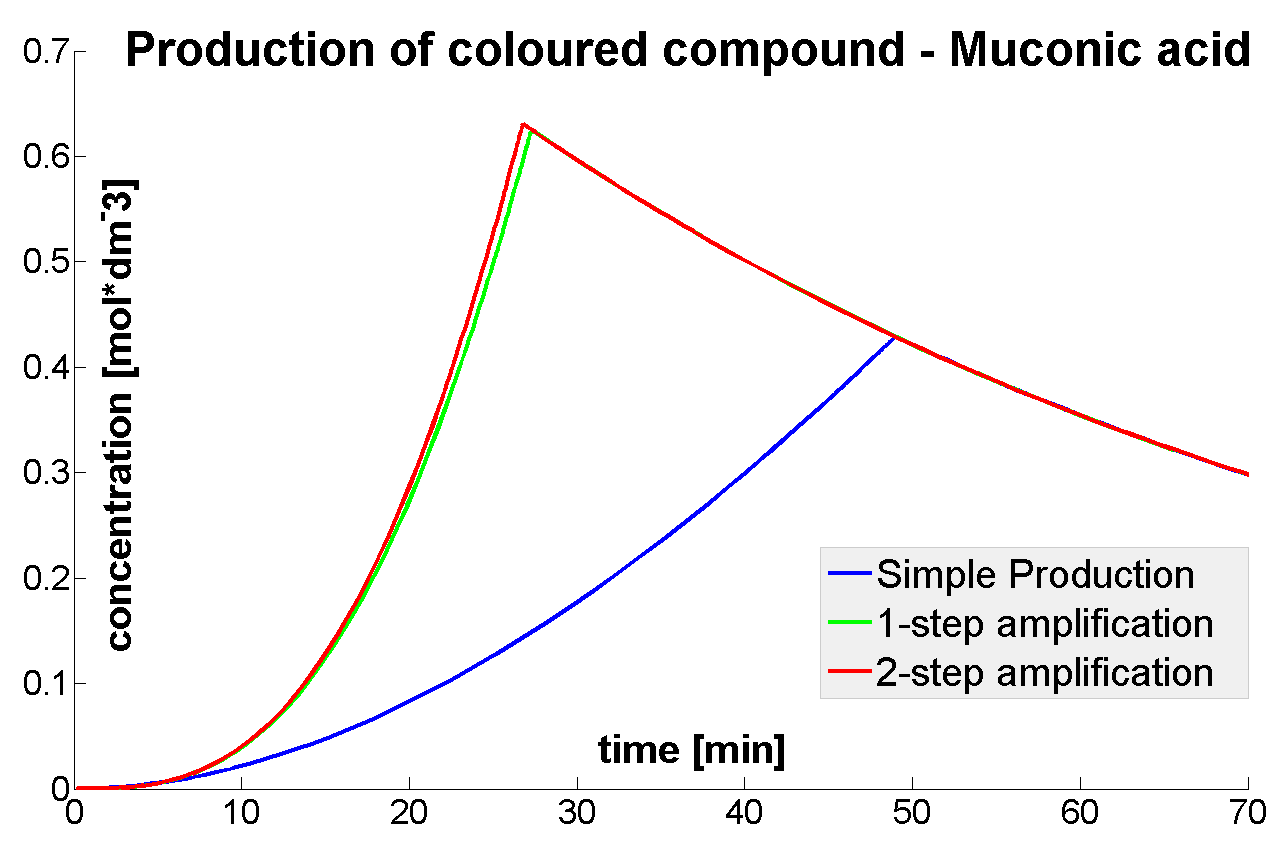
| - | |Diagram | + | |Diagram showing 4 models that have been considered. From top are shown: Simple production of arbitrary colour output by transcription and translation (<i>blue arrows</i>). Second diagram from top shows dioxygenase (C230) that acts on catechol (cat.) to produce muconic acid. Third diagram from top shows TEV protease activating inactive dioxygenase which acts on catechol to produce colour. The species that are shown in front of vertical line indicating beginning of experiment mean that they have been accumulated in the cell. The bottom most image introduces inactive split TEV protease attached to coiled-coil. Both inactive compounds have active site for TEV to act which results in multiple possibilities of action. |
|} | |} | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Major assumptions: | Major assumptions: | ||
Revision as of 23:15, 20 October 2010
| Temporary sub-menu: Output Amplification Model; Surface Protein Model; Wet-Dry Lab Interaction; Dry Lab Diary; |
| Introduction to modelling |
In the process of designing our construct two major questions arose which could be answered by computer modelling:
|
| Results & Conclusions | ||
Output Amplification Model
|
 "
"