Team:Lethbridge/Project
From 2010.igem.org
Liszabruder (Talk | contribs) |
Liszabruder (Talk | contribs) |
||
| Line 2: | Line 2: | ||
<html> | <html> | ||
| + | <br> | ||
<br> | <br> | ||
| Line 37: | Line 38: | ||
| - | + | <br> | |
<br> | <br> | ||
Liszabruder (Talk | contribs) |
Liszabruder (Talk | contribs) |
||
| Line 2: | Line 2: | ||
<html> | <html> | ||
| + | <br> | ||
<br> | <br> | ||
| Line 37: | Line 38: | ||
| - | + | <br> | |
<br> | <br> | ||
Overall project
Contents |
As a team from Alberta, we know of the economic benefit that the oils sands have brought to the province. Many members on our team have friends and family employed around the province in the energy industry, however, we are also aware of the environmental impact of the oils sands, specifically the tailings ponds. We feel it is important not only to clean the tailings ponds, but be able to extract the potential energy of the residue bitumen concurrently. Our team has previously worked on related projects in past years providing us with experience in techniques required to complete this project and potential problems that may arise. Also, being able to work on a project where we will be able to test our bacteria on real life samples gives us a physical goal as opposed to other, more esoteric projects.
The long term goal of our project will be to create a dry powder that will enzymatically degrade tailing ponds with no live bacteria, similar to how the pills taken for lactose intolerance contain enzymes for dealing with lactose. However this will require many intermediate steps. The initial goal of our project will be to create four separate modules for use in a bacteria test chassis. These modules will 1) degrade target contaminants into useful metabolites, 2) allow the bacteria to move towards areas of high chemical concentration, 3) induce DNA degradation and 4) concentrate the modules in microcompartments. Upon successful testing of each module, they will be combined into one cell chassis (E. coli). This chassis will then be optimized to live in a tailing pond environment by selective evolution. By combining our modules in microcompartments, we can extract the enzymes required to process tailings ponds and avoid putting live genetic organisms into the wild.
Using selective evolution in the presence of catechol we will engineer an E. coli which is able to live in high concentrations of catechol. This will allow for optimization of the catechol degradation pathway using. The next step will be to further use selective evolution with the tailings sample so that the pathway will be optimized for the environmental conditions that the pathway will administered to.
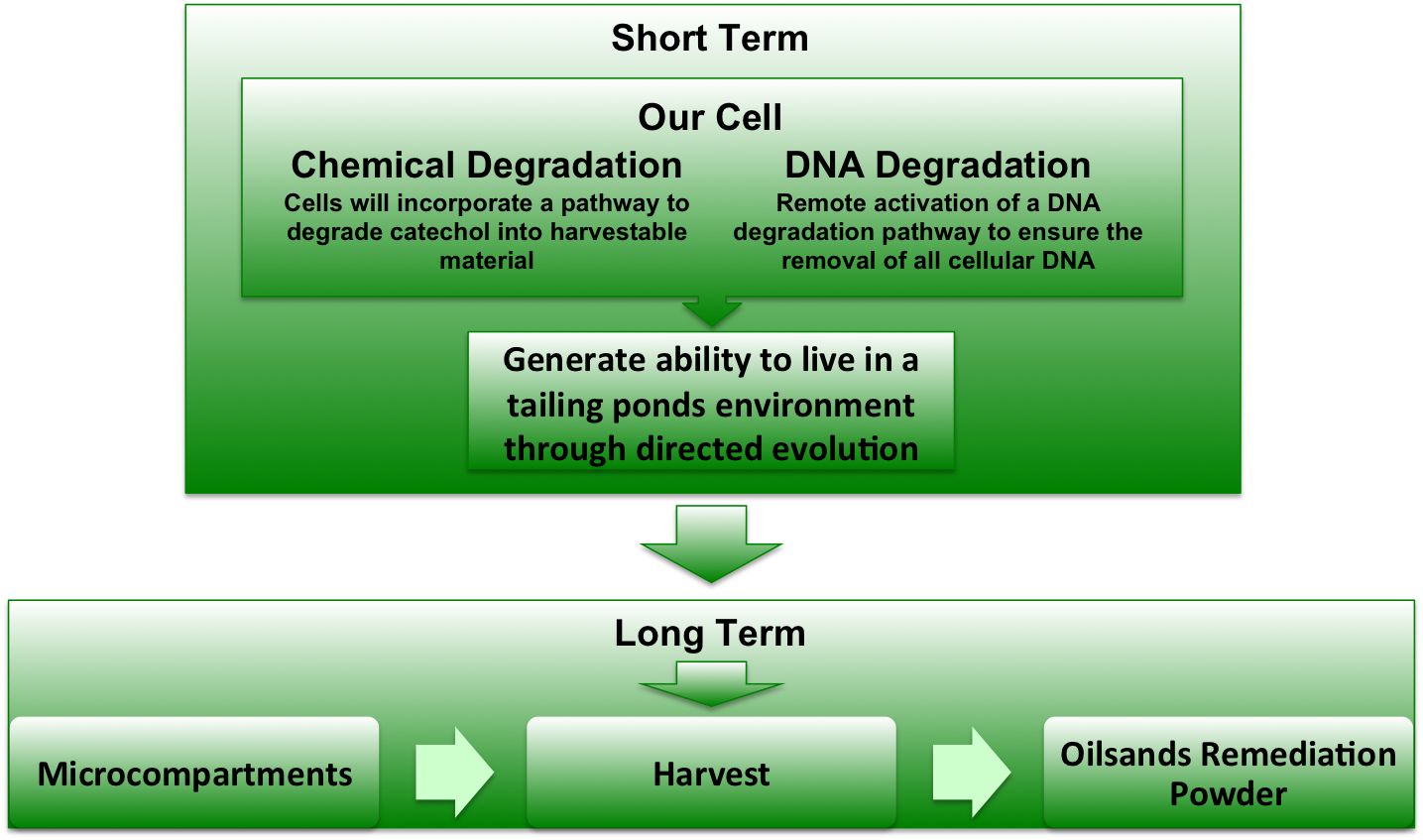 Figure 1. A depiction of the project overview.
Figure 1. A depiction of the project overview.
We expect to have the biobrick parts submitted to the registry for all of the separate modules. Once we have the biobrick parts we will then be able to assemble the proposed system of the catechol degradation pathway, chemotaxis, DNA degradation and microcompartments. We will characterize all the modules, but the catechol dioxygenase will be our priority. Our short-term goal will be to provide characterized biobricks for the degradation of catechol in tailings.
We will accomplish the optimization of the system by selective evolution. To accomplish this we will use the tailings sample provided to optimize the assembled of the optimized catechol degradation pathway.
We will fully assemble the optimized modules for the tailings sample by selective evolution in preparation for use. The optimized pathway will be created into an easily administered powder which can be added to the tailing ponds which will degrade the catechol without addition of live organisms. This will allow for the tailings’ cleanup which will leave a smaller environmental footprint while simultaneously decreasing the time necessary for the tailings cleanup.