Team:BCCS-Bristol
From 2010.igem.org
(Difference between revisions)
| (44 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:BCCS-Bristol/Header}} | {{:Team:BCCS-Bristol/Header}} | ||
| + | __NOTOC__ | ||
| + | <html><center> | ||
| + | <div style="width:64em;"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/b/bc/AgrEcoli_alt_logo.png" width="100%"/> | ||
| + | </div> | ||
| + | </center> | ||
| + | </html> | ||
| - | + | <center> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | '''agrEcoli is a device based on modified''' '''''E.coli''''' '''bacteria that detects and signals the presence of nitrates. This allows farmers to map the nutrient content of their fields and optimize their fertiliser use. For more information, see our [https://2010.igem.org/Team:BCCS-Bristol/Project Project Abstract]''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ==Achievements== | ||
| + | </center> | ||
| + | ===Wetlab=== | ||
| - | + | {| | |
| + | | | ||
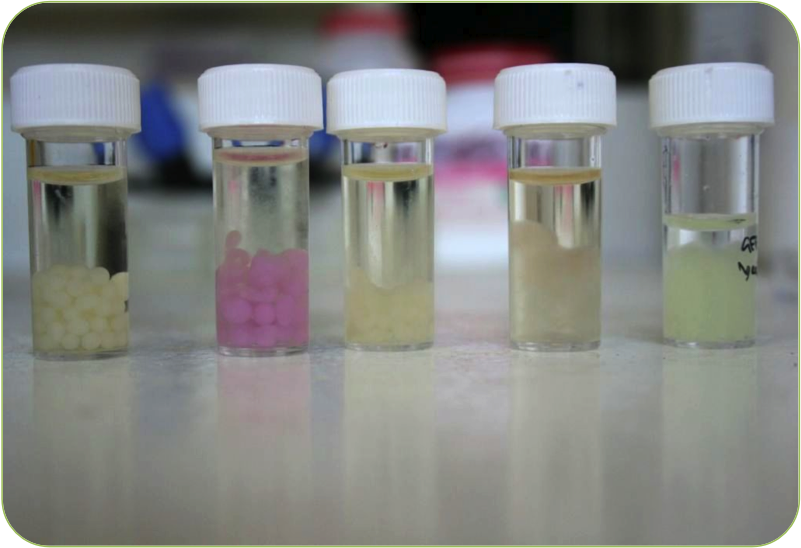
| - | + | * '''A Well Characterised New BioBrick''' | |
| + | :Our new BioBrick ([http://partsregistry.org/Part:BBa_K381001 BBa_K381001]) causes GFP expression in the presence of nitrates. | ||
| + | * '''Elegant Solution to Signal Calibration''' | ||
| + | :By using constitutive RFP expression as a baseline, we have found a reliable and accurate way of quantifying nitrate levels in soil. | ||
| + | * '''Novel Use of Cell Encapsulation''' | ||
| + | :By encapsulating our bacteria in gellan beads, we can keep our bacteria contained and concentrated. This improves visibility on soil, and enhances the environmental safety of our device. | ||
| + | * '''Characterising a Pre-Existing BioBrick''' | ||
| + | :To better inform our own work, and to add knowledge to the BioBrick Registry, we have characterised Edinburgh 2009’s PyeaR BioBrick ([http://partsregistry.org/Part:BBa_K216009 BBa_ K216009]). | ||
| + | |[[Image:Beads.png|thumbnail|400px|right|Our new beads]] | ||
| + | |} | ||
| + | {| | ||
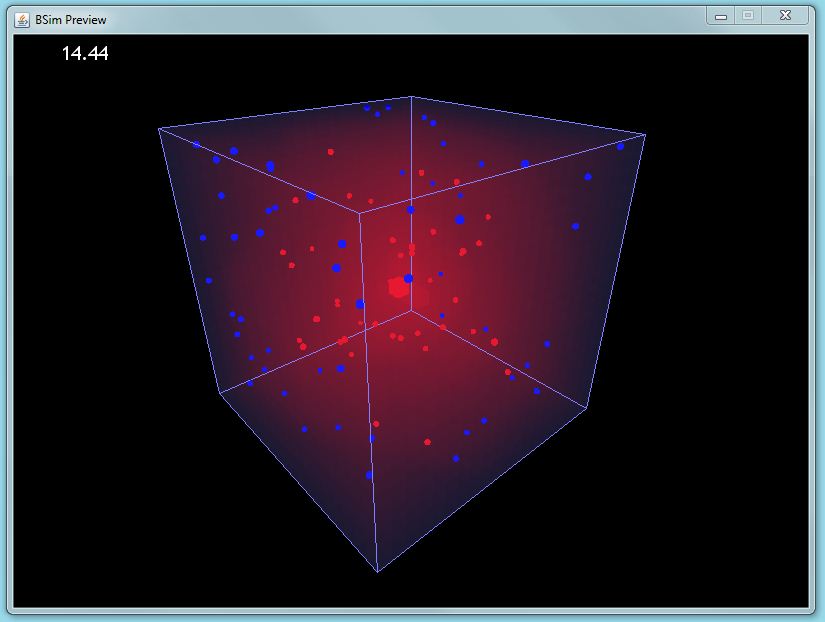
| + | |[[Image:GUI preview sample.JPG|thumbnail|400px|left|BSim screenshot]] | ||
| + | [[Image:Depth0.png|thumbnail|400px|left|Gel strand rendered in BSim]] | ||
| - | |||
| - | + | | | |
| - | + | ===Modelling=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | *'''BSim Environmental Interactions''' | ||
| - | + | :We have extended BSim, our agent-based modelling framework, to model interactions between bacteria and their environment. We have added 3-Dimensional mesh structures to our simulations, and added an adaptive chemical field routine that can solve partial differential equations in an arbitrary 3-D space without any risk of numerical instability. | |
| + | *'''BSim Graphical User Interface (GUI)''' | ||
| - | + | :We have made BSim more widely accessible by creating a user-friendly and intuitive graphical user interface. This makes BSim accessible to the entire synthetic biology community, rather than just those with JAVA programming knowledge. | |
| + | *'''Gene Regulatory Network Modelling''' | ||
| + | :We have investigated the behaviour of our bacteria by creating a mathematical model of their behaviour. This model could then be analysed using numerical and analytic methods. | ||
| + | *'''Collaboration''' | ||
| + | :Our modelling team have helped to simulate the UCL 2010 team’s new system. Working with another team has also helped to inform the design of the BSim GUI. | ||
| + | *'''agrEcoli Cost estimates''' | ||
| - | + | :In support of our human practices work, our modelling team have looked into the cost to farmers of using agrEcoli, and how much money and fertiliser they can expect to save. | |
| - | + | ||
| - | | | + | |} |
| - | | | + | |
| + | {| | ||
| | | | ||
| - | '' | + | ===Human Practices=== |
| - | |[[Image:BCCS- | + | |
| - | |- | + | *'''Publicising agrEcoli''' |
| - | + | ||
| - | + | :Our new approach to human practices, building on previous work by iGEM teams, is a publicity campaign. By presenting our prototype as a functioning and marketable product, we've framed a hypothetical situation in which our project could be released. | |
| + | |||
| + | *'''Public Engagement''' | ||
| + | |||
| + | :Our team has visited a school and spoken on two radio shows in order to communicate our ideas with the general public. | ||
| + | |||
| + | |[[Image:BCCS-Bristol-ClevedonVisit-03.jpg|thumbnail|400px|right|Public Engagement- Clevedon School Visit]] | ||
|} | |} | ||
| - | + | ||
| + | |||
| + | <center> | ||
| + | |||
| + | ==Sponsors== | ||
| + | [[Image:BCCS_Sponsors_Logo_Banner.png|upright= 3.3|frameless|center]] | ||
| + | </center> | ||
Latest revision as of 00:55, 28 October 2010
BCCS-Bristol
iGEM 2010
iGEM 2010

agrEcoli is a device based on modified E.coli bacteria that detects and signals the presence of nitrates. This allows farmers to map the nutrient content of their fields and optimize their fertiliser use. For more information, see our Project Abstract
Achievements
Wetlab
|
|
|
Modelling
|
Human Practices
|
 "
"