Team:TU Delft/5 August 2010 content
From 2010.igem.org
Contents |
Groningen Team
Today three members had an appointment with one of the professors at our department. We heard of this and invited them to come over to meet each other.
Lab Work
Alkane Sensing, Solvent Tolerance and Salt Tolerance
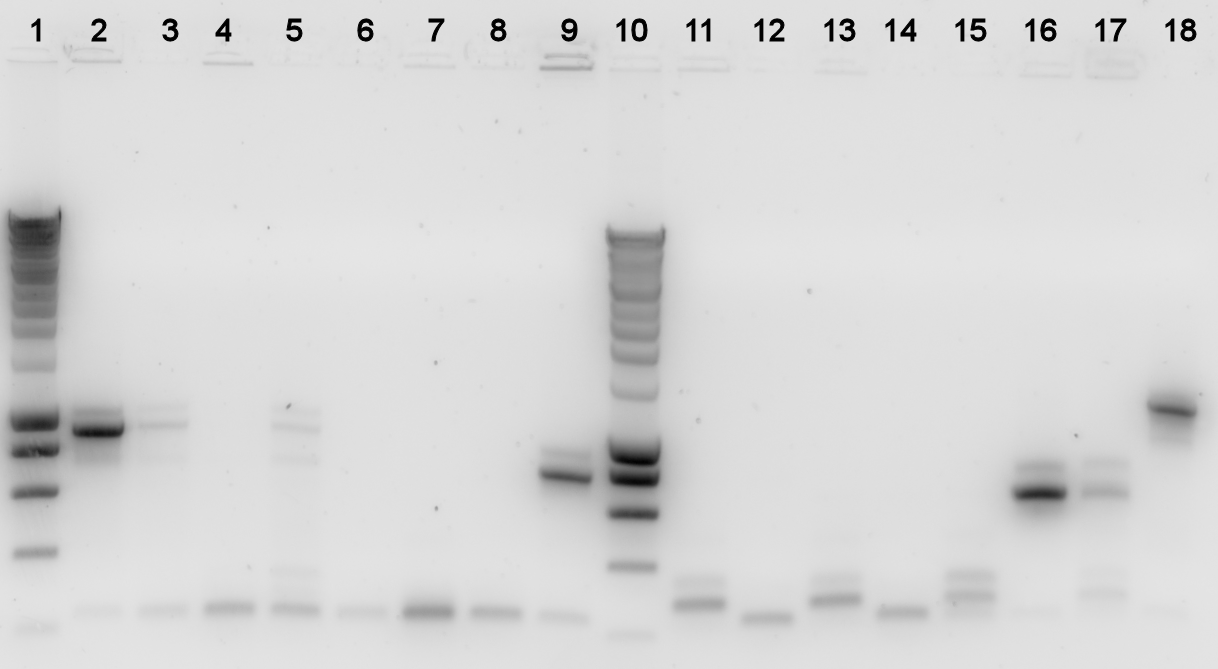
The plates containing yesterday's ligations contained colonies, to check whether they really contain the desired BioBrick a colony PCR was done, and the used colonies were grown in liquid LB medium over night. The results from the PCR were analysed on a 1% agarose gel.
Lane Description:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| 1 | SmartLadder | n/a | n/a | n/a | |
| 2 | BBa_K398101 | 929 | G00101 + G00100 | ✓ | |
| 3 | BBa_K398101 | 929 | G00101 + G00100 | ✗ | |
| 4 | BBa_K398101 | 929 | G00101 + G00100 | ✗ | |
| 5 | BBa_K398101 | 929 | G00101 + G00100 | ✗ | |
| 6 | BBa_K398101 | 929 | G00101 + G00100 | ✗ | |
| 7 | BBa_K398402 | 758 | G00101 + G00100 | ✗ | |
| 8 | BBa_K398402 | 758 | G00101 + G00100 | ✗ | |
| 9 | BBa_K398402 | 758 | G00101 + G00100 | ✓ | |
| 10 | SmartLadder | n/a | n/a | n/a | |
| 11 | BBa_K398402 | 758 | G00101 + G00100 | ✗ | |
| 12 | BBa_K398402 | 758 | G00101 + G00100 | ✗ | |
| 13 | BBa_K398403 | 656 | G00101 + G00100 | ✗ | |
| 14 | BBa_K398403 | 656 | G00101 + G00100 | ✗ | |
| 15 | BBa_K398403 | 656 | G00101 + G00100 | ✗ | |
| 16 | BBa_K398403 | 656 | G00101 + G00100 | ✓ | |
| 17 | BBa_K398403 | 656 | G00101 + G00100 | ✗ |
Alkane degradation
Colony PCR
The morning started with a gel checking the PCR products of the one BioBrick that didn't give positives yesterday. This gel showed one positive, but when I centrifuged the cells, they gave a red pellet. This indicated that the plasmid unfortunately was not the desired BioBrick, but the original destination plasmid, so we'll have to try again.
Lane description:
| # | Description | Primers | Expected length (bp) | ✓ | ✗ |
| L | SmartLadder | n/a | n/a | n/a | n/a |
| 1-9 | 020K | G00100 + G00101 | 2428 | 6 | rest |
Plasmid Isolation
Of the colonies which gave a positive PCR yesterday, the plasmids were isolated using a QIA-gen miniprep kit. The following concentrations were obtained:
| BioBrick | Composed of | Concentration (ng/μL) |
| 007A (1) | J61100-alkB2 | 33.8 |
| 007A (2) | J61100-alkB2 | 32.0 |
| 012K | J61100-rubR-B0015 | 42.7 |
| 021A | J61107-ALDH-B0015 | 46.3 |
| 021K (5) | J61107-ALDH-B0015 | 7.3 |
| 021K (1) | J61107-ALDH-B0015 | 11.1 |
| 021K (2) | J61107-ALDH-B0015 | 12.9 |
For some reason the 021K plasmid simply didn't want to be purified. All three of the isolates had a very low concentration. Next week we'll try another plasmid isolation method and see if that works.
Digestion
A few of the plasmids just isolated were digested:
| # | Sample | Enzyme 1 | Enzyme 2 | Enzyme 3 | Buffer | BSA | Needed fragment |
| 1 | 009A | EcoRI | SpeI | PvuI | 2 (Biolabs) | ✓ | ‘E - J61100 - rubA4 - S’ |
| 2 | 012K | EcoRI | XbaI | ✗ | 2 (Biolabs) | ✓ | ‘E – J61100 - rubR - B0015 – X’ |
| 3 | 007A | EcoRI | SpeI | PvuI | 2 (Biolabs) | ✓ | ‘E – J61100 - alkB2 – S’ |
| 4 | 008A | EcoRI | XbaI | ✗ | 2 (Biolabs) | ✓ | ‘E – J61100 - rubA3 – X’ |
The digestions were checked on a gel:
Lane description
| # | Description | Expected size (bp) | OK? | # | Description | Expected size (bp) | OK? |
| L | Smartladder (3μl) | n/a | n/a | 6 | 009A cut | 267 | ✓ |
| 1 | 007A cut | 1323 | ✓ | 6 | 009A uncut | n/a | |
| 2 | 007A uncut | n/a | 7 | 012K cut | 3552 | ✓ | |
| 3 | 008A cut | 2640 | ✓ | 8 | 012K uncut | n/a | |
| 4 | 008A uncut | n/a |
Ligation
Following the digestions, the fragments were [Team:TU_Delft/protocols/ligation|ligated]] for 4 hours.
| # | BioBrick | Fragment 1 | Recipient vector |
| 1 | 013AK | 15 μL ‘E – J61100 - rubA4 – S’ | 15 μL ‘X – J61100 - rubR - B0015 - pSB1AK3 – E’ |
| 2 | 011A | 15 μL ‘E – J61100 - alkB2 – S’ | 15 μL ‘X – J61100 - rubR - B0015 - pSB1AK3 – E’ |
Transformations
With the ligation mixes we transformed Top10 competent cells, and grew them overnight on solid LB plates.
 "
"