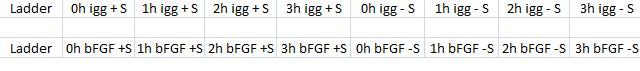Team:Stockholm/30 July 2010
From 2010.igem.org
Contents |
Nina
IPTG induction on IgG protease-troubleshooting
I have had trouble overexpressing IgG protease (ideS), thus I ran two coomassie gels on IPTG treated BL21 samples. The gels were prepared according to our SDS-PAGE mixtures under protocol.
The IPTG treatment was carried out as once before. As a positive control I treated bFGF in its original vector with IPTG, since I knew it has been overexpressed previously. In addition I had each sample in two versions, such as one with and one without sonication. This was to investigate if the results would have any effect of the sonication. However, I don't recommend not sonicating samples since I observed they get very viscous when loading on to gels.
Arrangement on gels:
Andreas
DNA concentration measurements
Continued from 17/7 samples.
DNA concentrations
| Sample | DNA conc (ng/μl) | A260/A280 |
| pSB1C3.BBa_J18930 A | 133.5 | 1.92 |
| pSB1C3.BBa_J18930 B | 37.84 | 1.95 |
| pSB1C3.BBa_J18931 A | 139.9 | 1.97 |
| pSB1C3.BBa_J18931 B | 51.21 | 2.04 |
| pSB1C3.BBa_J18932 A | 22.36 | 2.29 |
| pSB1C3.BBa_J18932 B | 79.83 | 1.95 |
| pSB1C3.SOD A | 120.6 | 2.02 |
| pSB1C3.SOD B | 40.32 | 1.95 |
| pSB1C3.yCCS A | 104.1 | 1.95 |
| pSB1C3.yCCS B | 86.70 | 1.98 |
Verification of yCCS site-directed mutagenesis
Site-directed mutagenesis performed by Mimmi, and colony PCR using pSBx VF2 and VR primers. Successful mutagenesis (i.e. removal of the unwanted EcoRI site) tested by digestion of colony PCR amplicons.
Samples (carried on pSB1C3)
| yCCS B1 | non-mutagenized controls |
| yCCS B2 | |
| yCCS A | mutagenized clones |
| yCCS B | |
| yCCS C | |
| yCCS D | |
| yCCS E | |
| Control | Blank |
Digestion
- 17 μl dH2O
- 2 μl 10X FastDigest Green buffer
- 10 μl plasmid DNA
- 1 μl FastDigest EcoRI
Incubation: 37 °C, 25 min
Gel verification
1 % agarose, 80 V, 50 min (?)
Loaded amounts:
- 2 μl GeneRuler 1 kb ladder
- 4 μl undigested sample
- 6 μl digested sample.
Expected bands
- Undigested samples: 1076 bp
- Digested samples:
- EcoRI removed: ≈120 bp, ≈955 bp
- EcoRI remaining: ≈120 bp, ≈705 bp, ≈250 bp
Results
≈700 bp bands clearly visible in non-mutagenized samples, while these are absent in the mutagenized samples. Also, weak bands are visible at around 250 bp for non-mutagenized samples.
Results indicate successful removal of the intragenous EcoRI site.
LMWP ligation/construction
LMWP primers diluted to 10 pmol/μl in 100 μl samples
Primer ligation
Ligation tubes
| N-LMWP | C-LMWP |
| 1 μl LMWP_N_F1 | 1 μl LMWP_C_F1 |
| 1 μl LMWP_CN_F2 | 1 μl LMWP_CN_F2 |
| 1 μl LMWP_CN_R1 | 1 μl LMWP_CN_R1 |
| 1 μl LMWP_N_R2 | 1 μl LMWP_C_R2 |
| 11 μl dH2O | 11 μl dH2O |
| 4 μl 5X Rapid Ligation buffer | 4 μl 5X Rapid Ligation buffer |
Incubation
- 95 °C - 10:00
- 60 °C - 5:00
- 37 °C - 5:00
- Temp. lowered to 22 °C, samples collected by centrifugation
- Added 1 μl Fermentas T4 ligase
- 22 °C - 15:00
- Cooled down on ice.
LMWP insertion
Insert = LMWP ligated from above
Insertion performed in two different vector:insert ratios:
- 1:10
- 1:100
| N-LMWP & C-LMWP | |
|---|---|
| 1:10 | 1:100 |
| 1 μl insert (0.5 pmol) | 10 μl insert (5.0 pmol) |
| 2 μl pSB1C3 (0.05 pmol) | 2 μl pSB1C3 (0.05 pmol)† |
| 4 μl 5X Rapid Ligation buffer | 4 μl 5X Rapid Ligation buffer |
| 12 μl dH2O | 3 μl dH2O |
| 1 μl T4 ligase | 1 μl T4 ligase |
†For C-LMWP 1:100, pSB1A3 vector was used instead of pSB1C3.
Incubation in 22 °C, 15 min.
Transformation of ligated vectors into Top10; transformation volume 3 μl. Transformed cells plated and grown on LB agar with appropriate antibiotic 37 °C ON. Ligation samples saved in -20 ° for possible gel verification.
ON cultures
Verified pSB1C3.yCCS A & B clones were inoculated in both 5 ml LB + Cm 25 and 3 ml LB Cm 25. Grown ON in 37 °C/250 rpm and 30 °C, respectively.
Four clones (A, B, C & D) were picked from Mimmi's mutagenized MITF plate and grown ON in 5 ml LB + Amp 100 in 37 °C, 250 rpm to attempt verification of site-directed mutagenesis. Also, two slightly red colonies were discovered on her plate. These were also picked (RED A & B) and grown under same conditions.
 |

|
 |

|
 |

|

|

|
 "
"