Team:Stockholm/2 August 2010
From 2010.igem.org
Contents |
Andreas
CPP DNA synthesis
Designed a CPP cluster to be sent for synthesis, containing all CPPs (with Freiburg prefixes and suffixes) as follows:
(ApaI)—C-Tra10—(BamHI)—N-Tra10—(ClaI)—C-TAT—(NcoI)—N-TAT—(SmaI)—C-LMWP—(BglII)—N-LMWP—(HindIII)
>CPP_cluster GGGCCCGCGAATTCGCGGCCGCTTCTAGATGGCCGGCGCGGGTTACCTGCTGGGTAAAAT CAACCTGAAAGCGCTGGCGGCGCTGGCGAAAAAAATCCTGACCGGTTAATACTAGTAGCG GCCGCTGCAGGCGGATCCGCGAATTCGCGGCCGCTTCTAGATGGCGGGTTACCTGCTGGG TAAAATCAACCTGAAAGCGCTGGCGGCGCTGGCGAAAAAAATCCTGACCGGTTAATACTA GTAGCGGCCGCTGCAGGCATCGATGCGAATTCGCGGCCGCTTCTAGATGGCCGGCTACGG TCGTAAAAAACGTCGTCAGCGTCGTCGTACCGGTTAATACTAGTAGCGGCCGCTGCAGGC CCATGGGCGAATTCGCGGCCGCTTCTAGATGTACGGTCGTAAAAAACGTCGTCAGCGTCG TCGTACCGGTTAATACTAGTAGCGGCCGCTGCAGGCCCCGGGGCGAATTCGCGGCCGCTT CTAGATGGCCGGCGTTTCTCGTCGTCGTCGTCGTCGTGGTGGTCGTCGTCGTCGTACCGG TTAATACTAGTAGCGGCCGCTGCAGGCAGATCTGCGAATTCGCGGCCGCTTCTAGATGGT TTCTCGTCGTCGTCGTCGTCGTGGTGGTCGTCGTCGTCGTACCGGTTAATACTAGTAGCG GCCGCTGCAGGCAAGCTT
Cm stocks
Prepared 4 x 1 ml 25 μg/μl Cm stocks.
LB agar plates
Prepared Cm and Amp LB agar plates as follows:
- 20x 25 μg/ml Cm
- 20x 100 μg/ml Amp
---
Mimmi
MITF - amplifying and moving the gene to pSB1C3
- Using AmplifX, suggested Ta1 = 54.8°C, Ta2 = 62.6°C
- Primers
- (MW/10)/(OD*33)=df
- 1000/df=V for 100µM
- 1:10=V for 10µM
- 1000/df=V for 100µM
- MITF_F: 70.59µl H2O --> 1:10
- MITF_R: 541.88µl H2O --> 1:10
| Mix | (µl) | X4 (µl) | Mix control | (µl) | X5 (µl) | contitions | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| H2O | 39.5 | 158 | H2O | 22.5 | 112.5 | time | °C | |||
| F primer | 0.75 | 3 | F primer | 1 | 5 | 2m | 94 | |||
| R primer | 0.75 | 3 | R primer | 1 | 5 | 30s | 94 | ) | ||
| buffer | 5 | 20 | DNA | 0.5 | 5*0.5 | 30s | 55 | > 5 cycles | ||
| Pfx pol | 0.5 | 2 | tot | 25 | 125 | 1m30s | 68 | ) | ||
| dNTPs | 1.5 | 6 | 30s | 94 | ) | |||||
| MgSO4 | 1 | 4 | 30s | 62 | > 25 cycles | |||||
| DNA | 1 | 4 | 1m30s | 68 | ) | |||||
| tot | 50 | 200 | 10m | 68 | ||||||
| oo | 10 | |||||||||
Nina
PCR on Tyrosinase
I ran a new PCR on the Tyrosinase since I realized that the band from the last PCR on this gene was incorrect (too high than expected). The gene Tyrosinase is about 1900 nt.
PCR Mix (100 ul):
- Fprimer 50 uM 1 ul
- Rprimer 50 uM 1 ul
- dNTP 10 mM 2 ul
- Buffer 5X 20 ul for phusion polymerase
- Buffer 10X 10 ul for fermenta (Pfu) polymerase
- phusion polymerase 1 ul
- fermenta (Pfu) polymerase 1 ul
- H2O 75 ul for phusion polymerase
- H2O 85 ul for fermenta (Pfu) polymerase
- DNA template 1 colony
Primer dilutions:
100 uM gets a 1:2 dilution to become 50 uM
cV = cV
100V = 50*10ul
V = 500 / 100 = 5 ul primer and fill up to 10 ul with H2O.
I aliquate the 100 ul in four PCR tubes, each with 25 ul PCR mix.
PCR prgm:
98 °C 2 min
2 * 29 cycles of:
- 98 °C 30 sec
- 50-72 °C 30 sec
- 72 °C 2 min
72 °C 1 min
4 °C ∞
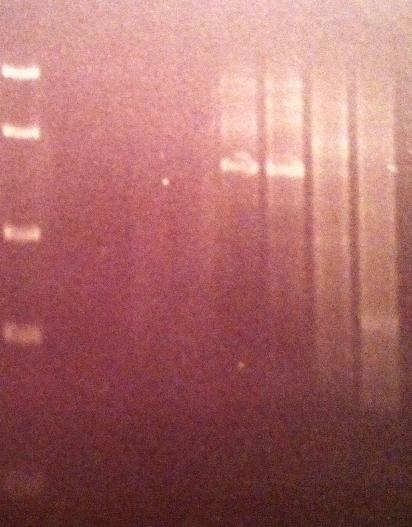
Agarose gel on Tyrosinase
I ran a 1 % agarose gel on the tyrosinase PCR productes.
Ladder: FastRuler™ Middle Range DNA Ladder, ready-to-use, 100-5000 bp Fermentas
Arrangement on gels:
1 = 50 °C - 4 = 72 °C
P1 and 2 look like they have succesfully PCR:ed tyrosinase.
Sequencing yCCS
I send two samples of yCCS for sequencing.
- 15 ul vector
- 1.5 ul 10uM VF2 primer
A: ASB0045 246
B: ASB0045 245
 |

|
 |

|
 |

|

|

|
 "
"