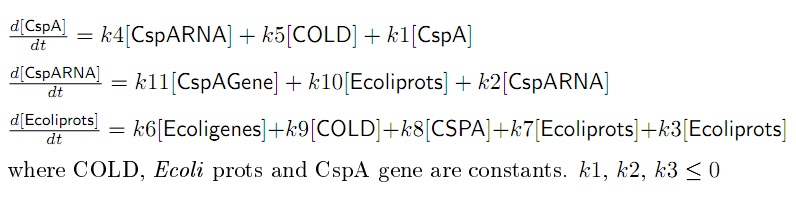Team:Mexico-UNAM-CINVESTAV/Modeling/Theory
From 2010.igem.org
Introduction to the model
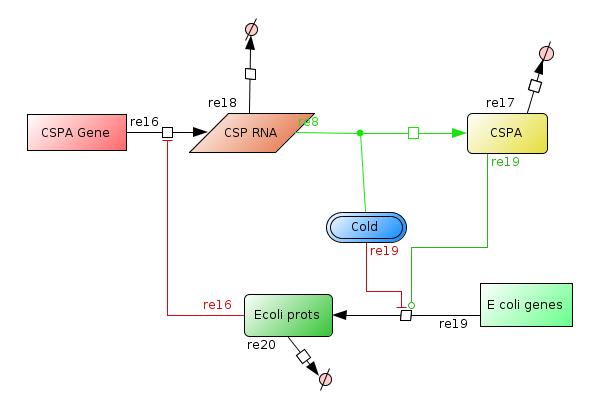
We designed a model of the Cold shock reponse system in bacteria by describing each possible interaction between the cold shock proteins (CSP), the cold induced proteins (CIP) and the non-cold induced proteins (non-CIP's) in Escherichia coli. (see Figure 1)
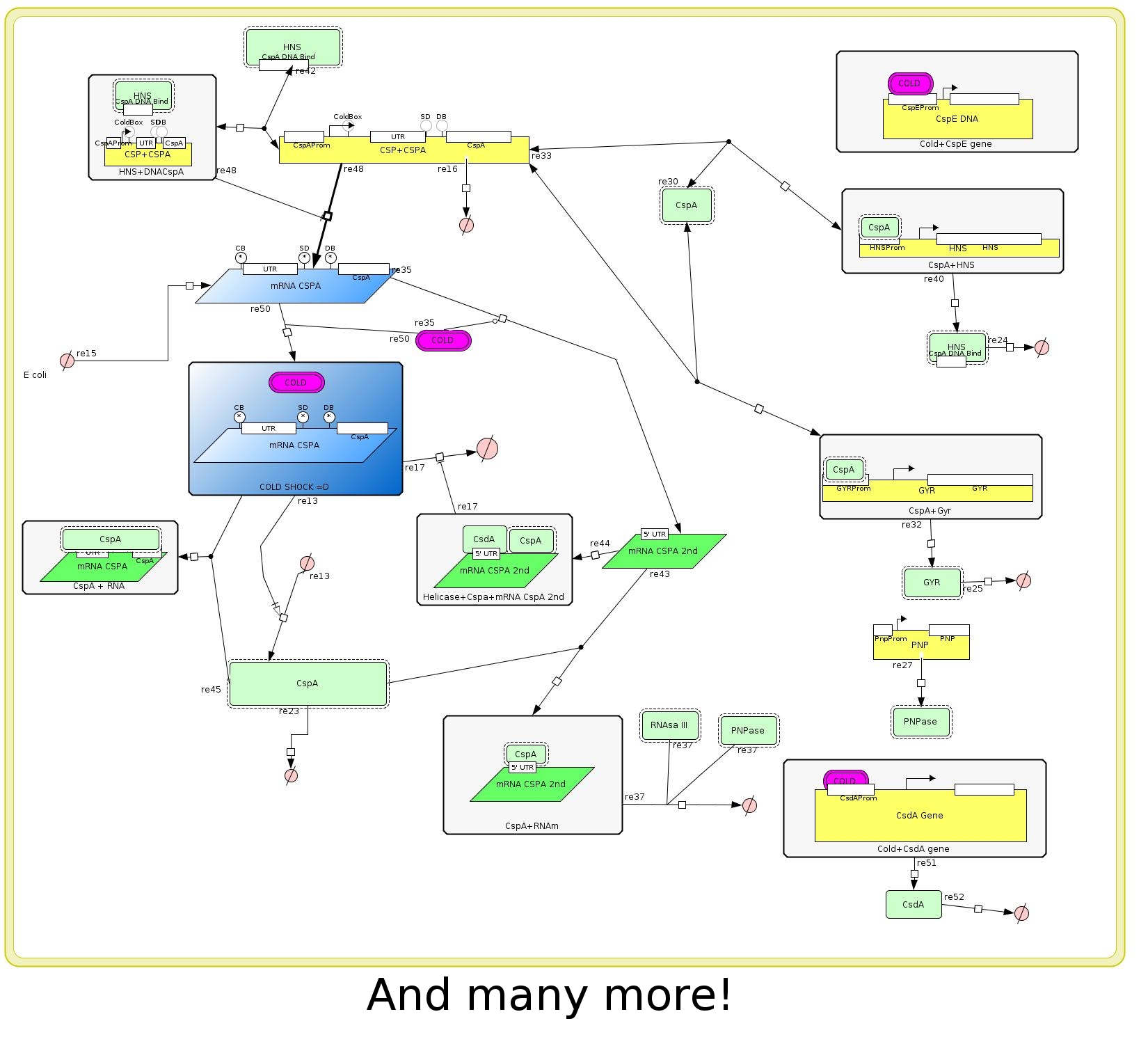
After a exhaustive research in the literature we realized that the amount of interaction among genes, mRNAS, proteins, enzymes, etc, was very high. Then we tried to describe only the more significant behaivor of cold shock response system with special attention to the major cold shock protein: Cold shock protein A (CspA) (Figure 2).
One of the goals of our project is isolate the regulation mechanism of the CspA gene to be able to control the expression of any gene. We realized that this mechanisms is very complex too because there are many levels of regulation transcriptional, post-transcriptional, mRNA stability and traductional. The amount of interactions was very high again so we tried to reduce our description to the main characteristics of the CspA regulation system. (Figure 3)
The model
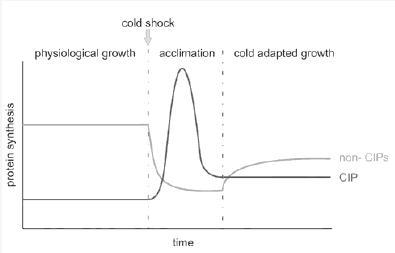
- It has been observed that the temperature stability affects mainly the translation of many E. coli proteins. And upon cold shock the properties of the CspA mRNA's gives them the possibility to be translated much better than the rest of the mRNA of E coli. The effect of the cold shock is represented with the species COLD, this species is constant over time and means that the cell has had a temperature downshift from 37º to 10-15ºC.
- The CspA gene is represented by the species CSPA GENE, which transcribe CSPA RNA at high level when COLD. Later the translation of the gene to CSP proteins is mediated by COLD and it can be high enough only in low temperatures.
- CspA gene will act as an RNA chaperone to help the production of the Ecoli prots species.
- The E. coli genes is represented by the species E coli genes that is constant over time too, and led to the production of Ecoli prots species. This synthesis is affected when COLD is "present" and by the CSPA protein (RNA chaperone).
- Later the Ecoliprots species is expected to reach certain level that is enough to inactivate the production of the CSPA RNA at transciption.
- Our more importat assumptions are that the effect of low temperature, 10 to 15ºC, is represented by the species COLD. This species more than a physical effect represent the interaction of the "molecule COLD" with the system.
The other one is that the model only represents the system in cold shock enviroentment to the aclimatation phase. (See Figure 1 footer)
- The final set of equations that represent the system is in Figure 4. All the parameters are positive unless mentioned.
Collaboration with UNAM-Genomics team
We designed a model for the UNAM-Genomics team where we described the dynamics of three genes that are in a positive feedback cycle. This model is explicated in detail in our Collaboration section.
References
- Ermolenko, D.N., Makhatadze, G.I. (2002) Bacterial cold-shock proteins. Cell Mol. Life Sci. 59, 1902–1913.
- Horn G, Hofweber R, Kremer W, Kalbitzer HR (2007) Structure and function of bacterial cold shock proteins. Cell Mol Life Sci 64: 1457–1470
- Funahashi, A., Tanimura, N., Morohashi, M., and Kitano, H., CellDesigner: a process diagram editor for gene-regulatory and biochemical networks, BIOSILICO, 1:159-162, 2003 [doi:10.1016/S1478-5382(03)02370-9]
- Funahashi, A.; Matsuoka, Y.; Jouraku, A.; Morohashi, M.; Kikuchi, N.; Kitano, H."CellDesigner 3.5: A Versatile Modeling Tool for Biochemical Networks" Proceedings of the IEEE Volume 96, Issue 8, Aug. 2008 Page(s):1254 - 1265 [doi 10.1109/JPROC.2008.925458]
- Thieringer, H. A., Jones, P. G. and Inouye, M. (1998), Cold shock and adaptation. BioEssays, 20: 49–57. doi: 10.1002/(SICI)1521-1878(199801)20:1<49::AID-BIES8>3.0.CO;2-N
 |
 |
 |
 |
 "
"