Team:TU Delft/Project/sensing/results
From 2010.igem.org
(→BBa_K398326 pCaiF in numbers) |
(→CONCLUSIONS) |
||
| Line 57: | Line 57: | ||
=====CONCLUSIONS===== | =====CONCLUSIONS===== | ||
| - | + | The simple pCaiF promoter with crp-cAMP binding site has shown activity under limiting nutrient conditions. Therefore, '''this promoter can be used to enable catabolic shifts''' from glucose to '''new degradation pathways'''. | |
| + | |||
| + | From our point of view, this part will be very useful for future teams. Please also check the results obtained for the model developed for this part: | ||
| - | |||
| - | |||
[HTTP://2010.IGEM.ORG/TEAM:TU_DELFT#PAGE=MODELING/PCAIF-MODEL IN SILICO WORK] | [HTTP://2010.IGEM.ORG/TEAM:TU_DELFT#PAGE=MODELING/PCAIF-MODEL IN SILICO WORK] | ||
Revision as of 20:42, 27 October 2010

Sensing Results
pCaiF strength
[http://partsregistry.org/Part:BBa_K398326 BBa_K398326 pCaiF]
[http://partsregistry.org/Part:BBa_K398326 BBa_K398326 (pCaiF)] is by far the smallest part of our project. This part enable the expression of proteins at low glucose concentrations in order to mimic a diauxic shift for the alkane degradation system: Once the preferred substrate, glucose becomes limiting, the expression of alkane degradation genes under pCaiF control will enable the cells to shift from glucose metabolism to alkane degradation. The whole regulation is performed by a piece of just 51 bp.
The pCaiF regulation mechanism is really simple, [http://partsregistry.org/Part:BBa_K398326 BBa_K398326 (pCaiF)] contains a cAMP-crp complex binding domain, cAMP-crp is known as transcriptional regulator. When the glucose concentration in the environment is high, cAMP levels are low because there is a lot of energy source that can be metabolized by the cells. However during starvation periods cAMP levels increase and thus the concentration of the complex cAMP-crp that is known to activate at least 180 genes related to starvation response. Among those pCaiF a protein used during Carnitine anaerobic metabolism.
An organism expressing our proteins using [http://partsregistry.org/Part:BBa_K398326 BBa_K398326 (pCaiF)] could not be constructed. Nevertheless, we studied the regulatory Biobrick [http://partsregistry.org/Part:BBa_K398326 BBa_K398326] using [http://partsregistry.org/Part:BBa_E0240 BBa_E0240] in order to measure the output given by our part.
LB medium
First we used a rich medium (LB) and we diluted it with M9 medium without glucose (50% v/v) in order to show the differences in protein expression when the carbon source concentration is lower in a rich medium.
Our findings suggest that there is GFP production when the LB medium is diluted 50%. However the fluorescence signal produced was weak, moreover we didn't see an increase of GFP production during the stationary phase (starvation). This could be due to the fact that LB medium can keep the cAMP levels low for long periods of time.
M9 minimal medium
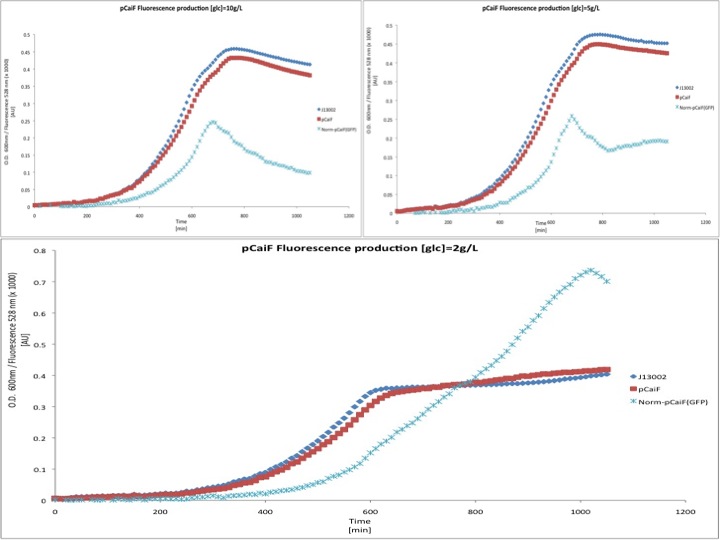
We then tested the response of pCaiF using minimal M9 medium at glucose concentrations of 10 g/L, 5 g/L and 2 g/L. From this experiment we expected to see a limitation of carbon source leading to a reponse by pCaiF.
In this experiment we clearly saw a significant increase in GFP production when the glucose initial concentration was 2 g/L compared to our result for an initial glucose concentration of 10 g/L; whereas at 5 g/L we found a slight increase in GFP production at the end of our measurements. From these plots we can conclude that our part is sensitive to cAMP levels as we expected. Moreover, we found that our part is active under carbon limitation conditions; which in our case it is detected at the beginning of stationary phase at low initial glucose concentrations, see figure.
Diauxic shift
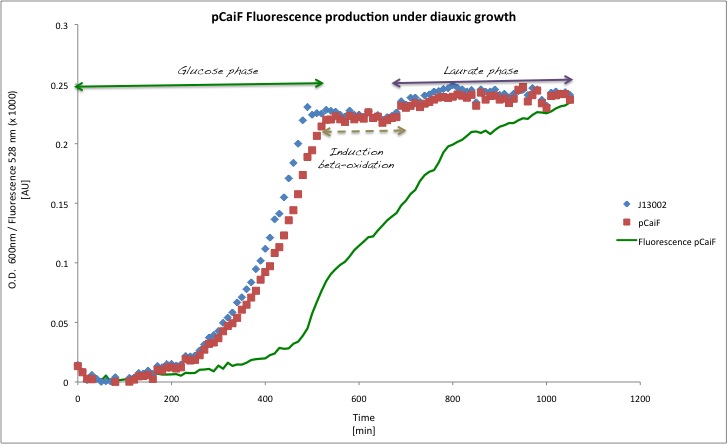
In order to see the effect of a second carbon source in the medium, we decided to decrease the initial glucose amount to 1 g/L and we added Potassium Laurate at a final concentration of 5 mM (secondary carbon source). This experiment is very close to the later application as lauric acid is metabolized via beta oxidation. We expect to observe a diauxic shift with high GFP expression during the lag phase of the metabolic switch.
Our findings in this experiment were really interesting, (1) the GFP signal first increases, indicating a limitation due to glucose depletion and (2) the GFP production decreases again when the catabolism of the second carbon source (lauric acid) starts. There is a clear change in the slope for the GFP profile over time with a max at about 8h.
Does [http://partsregistry.org/Part:BBa_K398326 pCaiF] really works under starvation conditions???
We had some doubts about the activity of pCaiF in the stationary phase because we usually observed a leaky production of GFP that followed the biomass profile. Therefore we decided to compare the GFP production during the exponential and stationary phase.
According to our results, there is indeed a difference between both conditions. Which is specially clear when the glucose initial concentration is 2 g/L. For the Laurate enriched medium (diauxic shift) we observed a remaining activity from the glucose limitation period and then a clear reduction during Laurate consumption.
[http://partsregistry.org/Part:BBa_K398326 BBa_K398326 pCaiF] in numbers
In order to convert Fluorescence arbitrary units to something meaningful, we used the parameters suggest in the part registry for [http://partsregistry.org/Part:BBa_E0040 E0040]. We divided the fluorescence random units by 79.429 (conversion factor to nM), then we calculated back the number of moles per well knowing that our reaction volume was 100 uL. If we multiply the total amount of moles per well by the [http://en.wikipedia.org/wiki/Avogadro_constant Avogadro's constant], we will get the total amount of GFP molecules produced in each data point. Then we applied a linear regression to each growth phase and we divided by the O.D. at 600nm in order to normalize the result ([http://partsregistry.org/RiPS RiPS]/O.D.).
| Condition | Exponential phase [GFP molecules/O.D.] | Stationary phase [GFP molecules/O.D.] |
|---|---|---|
| LB | 1.2516E+07 | -8.6245E+05 |
| 0.5LB | 1.8101E+07 | -2.5945E+05 |
| [glc]=10g/L | 6.6456E+06 | -6.7109E+06 |
| [glc]=5g/L | 8.3673E+06 | 4.2339E+06 |
| [glc]=2g/L | 7.2869E+06 | 3.9755E+07 |
| Diauxic growth (GLUCOSE PHASE) | 7.4517E+06 | 2.1543E+07 |
| DIAUXIC GROWTH (LAURATE PHASE) | 1.1435E+07 | 6.6428E+06 |
NOTE: Check at the part registry the definition of [http://partsregistry.org/RiPS RiPS]
CONCLUSIONS
The simple pCaiF promoter with crp-cAMP binding site has shown activity under limiting nutrient conditions. Therefore, this promoter can be used to enable catabolic shifts from glucose to new degradation pathways.
From our point of view, this part will be very useful for future teams. Please also check the results obtained for the model developed for this part:
[HTTP://2010.IGEM.ORG/TEAM:TU_DELFT#PAGE=MODELING/PCAIF-MODEL IN SILICO WORK]
WORK FOR NEXT TEAM
WE WOULD HAVE LIKED TO EXPRESS OUR RESULTS ACCORDING TO THE PROTOCOLS IN [HTTP://PARTSREGISTRY.ORG/POPS POLYMERASE PER SECOND]. THEREFORE THE OUTPUT OF B0032 AND A STANDARD PROMOTER GROWING IN M9 MEDIUM HAS TO BE MEASURED.

WORK FOR NEXT TEAM
WE WOULD HAVE LIKED TO EXPRESS OUR RESULTS ACCORDING TO THE PROTOCOLS IN [HTTP://PARTSREGISTRY.ORG/POPS POLYMERASE PER SECOND]. THEREFORE THE OUTPUT OF B0032 AND A STANDARD PROMOTER GROWING IN M9 MEDIUM HAS TO BE MEASURED.

 "
"