Team:TU Delft/Project/alkane-degradation/parts
From 2010.igem.org

Alkane Degradation Parts
Introduction
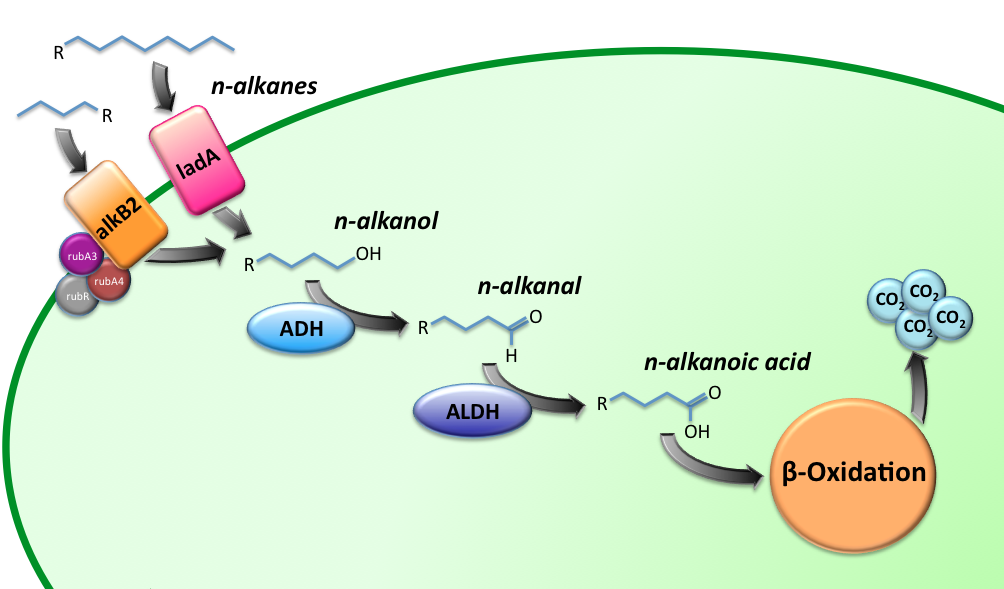
The Alkanivore E.coli strain has been designed to carry the genes required for the conversion of medium (C5-C13) and long chain (C15-C36) alkanes. A general scheme for the oxidation of the alkanes is illustrated in figure 1.
To create the alkane degradation constructs a number of genes encoding for alkane degradation enzymes were synthesized and combined with promoters and ribosome binding sites obtained from the BioBrick distribution plates. Combination of these genes resulted in the following BioBrick constructs (the intermediates have also been submitted to the registry).
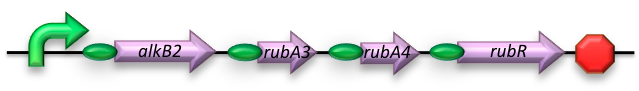
BBa_K398014 - Medium-chain (C5-C13) alkane conversion (alkB)
Our medium-chain hydrocarbon degrading strain contains the alkane hydroxylase (AH) native to Gordonia sp. TF6 [1]. This system facilitates the initial oxidation step of C5-C13 alkanes along with that of C5-C8 cycloalkanes towards their respective alcohols. Based on the literature on this topic [5] it is expected that the in-house mechanism of E.coli will be able to further degrade the products of this pathway.
The AH construct consists of the sequences encoding for:
- Alkane 1-monooxygenase (alkB2); an integral cytoplasmic membrane monooxygenase of which homologs have been reported for varying genus and species. This is the catalytic component of the AH system and as such oxidizes (cyclo)alkanes to their respective (cyclo)alkanols by transferring one oxygen atom from molecular oxygen to the alkane.
- Rubredoxin reductase (rubB); catalyzes the reduction of the second oxygen atom released from molecular oxygen using electrons supplied by NADH.
- Rubredoxin (rubA3); facilitates the transfer of electrons from NADH to rubredoxin reductase.
- Rubredoxin (rubA4); an electron-carrier protein required by the AH system.
The AH construct was designed to harbor all four of the required coding sequences -each behind its own RBS region- sharing a constitutive promoter.
View this part in the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398014 parts registry]
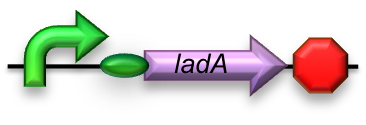
BBa_K398017 - Long-chain (C15-C36) alkane conversion (ladA)
For the first step in the long-chain alkane degradation pathway ladA was implemented [2]; a flavoprotein alkane monooxygenase native to Geobacillus thermodinitrificans NG-80-2. It has been found to specifically oxidize the terminal regions of alkanes ranging from C15 up to at least C36. The product is the corresponding primary alkanol. LadA forms a catalytic complex with flavin mononucleotide (FMN) which utilizes dioxygen to insert an oxygen atom into the substrate.
The general catalytic function involves three chemical processes:
- Reduction of the cofactor flavin mononucleotide (FMN to FMNH2) by NAD(P)H
- Reaction of FMNH2 with O2
- Binding, orienting, and activating the substrate for its oxygenation
LadA's ability to preferentially capture long-chain alkanes for oxidation sets it apart from other flavoprotein monooxygenases.
View this part in the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398017 parts registry]
BBa_K398018 - Medium-chain alkanol conversion (ADH)
The following step in the oxidation pathway is catalyzed by a zinc-independent alcohol dehydrogenase from Geobacillus thermoleovorans B23; a thermophilic bacterium [3]. The enzyme converts medium-chain alkanols into their respective alkanals by reduction of NAD into NADH.
View this part in the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398018 parts registry]
BBa_K398029 and BBa_K398030 - Medium-chain alkanal conversion (ALDH)
For the final step in the medium-chain oxidation the aldehyde dehydrogenase from the thermophile Geobacillus thermoleovorans B23 is implemented. It functions as an octamer, requiring NAD+ as coenzyme. The optimum condition for activity lies at temperatures between 50 and 55 degrees Celsius and a pH of 10 [4].
View BBa_K398029 in the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398029 parts registry]
View BBa_K398030 in the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398030 parts registry]
References
- Fujii, T., Narikawa, T., Takeda, K., Kato, J., Biotransformation of various alkanes using the Escherichia coli expressing an alkane hydroxylase system from Gordonia sp. TF6. Bioscience, biotechnology, and biochemistry, 68(10) 2171-2177 (2004)
- Liu Li, Xueqian Liu, Wen Yang, Feng Xu, Wei Wang, Lu Feng, Mark Bartlam, Lei Wang and Zihe Rao. Crystal Structure of Long-Chain Alkane Monooxygenase (LadA) in Complex with Coenzyme FMN: Unveiling the Long-Chain Alkane Hydroxylase. Journal of molecular biology, 376: 453-465 (2008)
- Tomohisa Kato, Asuka Miyanaga, Mitsuru Haruki, Tadayuki Imanaka, Masaaki Morikawa & Shigenori Kanaya. Gene Cloning of an Alcohol Dehydrogenase from Thermophilic Alkane-Degrading Bacillus thermoleovorans B23. Journal of Bioscience and Bioengineering 91(1):100-102 (2001)
- Tomohisa Kato, Asuka Miyanaga, Shigenori Kanaya, Masaaki Morikawa. Gene cloning and characterization of an aldehyde dehydrogenase from long-chain alkane-degrading Geobacillus thermoleovorans B23. Extremophiles 14:33-39 (2010)
- Sulzenbacher, G., et al., Crystal structure of E-coli alcohol dehydrogenase YqhD: Evidence of a covalently modified NADP coenzyme. Journal of Molecular Biology 342(2):489-502 (2004)
- http://mbel.kaist.ac.kr/lab/research/protein_en1.html
- Hoffmann F. and Rinas U. Stress Induced by Recombinant Protein Production in Escherichia coli Advances in Biochemical Engineering/Biotechnology, Vol. 89, pp. 73-92.(2004)

 "
"