Team:TU Delft/Project/alkane-degradation/parts
From 2010.igem.org
(→BBa_K398014 - Short-chain (C5-13) Alkane degradation) |
(→BBa_K398014 - Short-chain (C5-13) Alkane degradation) |
||
| Line 8: | Line 8: | ||
==BBa_K398014 - Short-chain (C5-13) Alkane degradation== | ==BBa_K398014 - Short-chain (C5-13) Alkane degradation== | ||
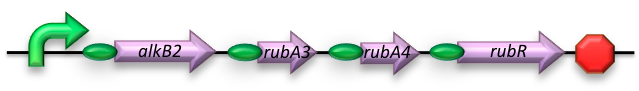
| - | The short-chain hydrocarbon degrading BioBrick contains the AlkB gene cluster from ‘’Gordonia sp. TF6’’[1]. This will facilitate the initial step of the oxidation of C5-13 alkanes as well as that of C5-8 cycloalkanes. It is expected that the in-house mechanism of E.coli will be able to further degrade the products of this pathway. The gene cluster is formed by the genes encoding for alkB2 (alkane 1-monooxygenase),rubA3 (rubredoxin), rubA4 (rubredoxin) and rubR (rubredoxin reductase). AlkB2 is a non-haem diiron monooxygenase membrane protein, reported for several genus and species. This monooxygenase oxidizes n-alkanes to the respective n-alkanols and requires three soluble electron-transfer proteins, rubredoxin (rubA3 & rubA4) and rubredoxin reductase (rubR). | + | The short-chain hydrocarbon degrading BioBrick contains the AlkB gene cluster from ‘’Gordonia sp. TF6’’[1]. This will facilitate the initial step of the oxidation of C5-13 alkanes as well as that of C5-8 cycloalkanes. It is expected that the in-house mechanism of E.coli will be able to further degrade the products of this pathway. The gene cluster is formed by the genes encoding for alkB2 (alkane 1-monooxygenase), rubA3 (rubredoxin), rubA4 (rubredoxin) and rubR (rubredoxin reductase). AlkB2 is a non-haem diiron monooxygenase membrane protein, reported for several genus and species. This monooxygenase oxidizes n-alkanes to the respective n-alkanols and requires three soluble electron-transfer proteins, rubredoxin (rubA3 & rubA4) and rubredoxin reductase (rubR). |
View this part in the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398014 '''parts registry'''] | View this part in the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398014 '''parts registry'''] | ||
Revision as of 13:33, 15 October 2010
Contents |
Alkane degradation parts
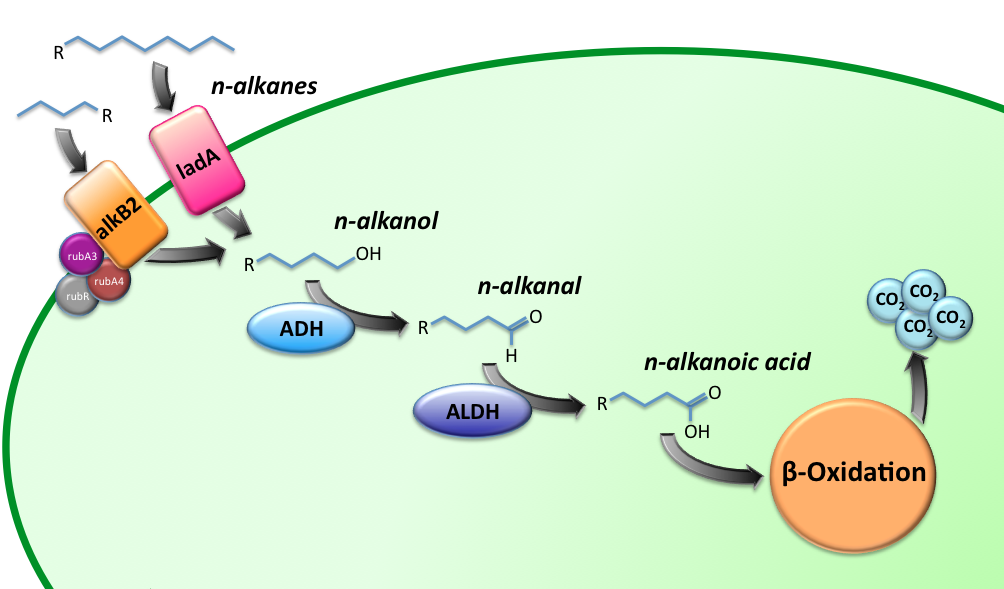
The Alkanivore E.coli strain has been designed to carry the genes required for the conversion of medium (C5-C13) and long chain (C15-C36) alkanes. A general scheme for the oxidation of the alkanes is illustrated in figure 1.
To create the alkane degradation constructs a number of genes encoding for alkane degradation enzymes were synthesized and combined with promoters and RBSs obtained from the BioBrick distribution plates. Combination of these genes resulted in the following BioBrick constructs (the intermediates have also been submitted to the registry).
BBa_K398014 - Short-chain (C5-13) Alkane degradation
The short-chain hydrocarbon degrading BioBrick contains the AlkB gene cluster from ‘’Gordonia sp. TF6’’[1]. This will facilitate the initial step of the oxidation of C5-13 alkanes as well as that of C5-8 cycloalkanes. It is expected that the in-house mechanism of E.coli will be able to further degrade the products of this pathway. The gene cluster is formed by the genes encoding for alkB2 (alkane 1-monooxygenase), rubA3 (rubredoxin), rubA4 (rubredoxin) and rubR (rubredoxin reductase). AlkB2 is a non-haem diiron monooxygenase membrane protein, reported for several genus and species. This monooxygenase oxidizes n-alkanes to the respective n-alkanols and requires three soluble electron-transfer proteins, rubredoxin (rubA3 & rubA4) and rubredoxin reductase (rubR).
View this part in the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398014 parts registry]
BBa_K398022 - Long-chain (C15-36) Alkane degradation
The long-chain degrading BioBrick contains three genes, forming a degradation pathway form alkanes to alkanoic acids. For the first step in this pathway ladA will be used [2], a long-chain alkane monooxygenase from ‘’Geobacillus thermodenitrificans’’ NG80-2. This gene facilitates the conversion of long-chain alkanes (up to at least C36) to their corresponding primary alcohols. Because of the length of the alkanes (and thus also of the pathway intermediates) the BioBrick will also contain an additional ADH and ALDH Bacillus thermoleovorans B23[3-4]. These genes will facilitate the conversion of these long-chain intermediates (alkanols & alkanals).
Unfortunately we weren't able to to get the three genes in one BioBrick, but because the genes can also work separately this wasn't a problem. We characterized the three genes separately.
View these parts in the parts registry:
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398017 ladA (BBa_K398017)]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398018 ADH (BBa_K398018)]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398019 ALDH (BBa_K398019)]
References
[1] Fujii, T., Narikawa, T., Takeda, K., Kato, J., Biotransformation of various alkanes using the Escherichia coli expressing an alkane hydroxylase system from Gordonia sp. TF6. Bioscience, biotechnology, and biochemistry, 68(10) 2171-2177 (2004)
[2] Liu Li, Xueqian Liu, Wen Yang, Feng Xu, Wei Wang, Lu Feng, Mark Bartlam, Lei Wang and Zihe Rao. Crystal Structure of Long-Chain Alkane Monooxygenase (LadA) in Complex with Coenzyme FMN: Unveiling the Long-Chain Alkane Hydroxylase. Journal of molecular biology, 376: 453–465 (2008)
[3] Tomohisa Kato, Asuka Miyanaga, Mitsuru Haruki, Tadayuki Imanaka, Masaaki Morikawa & Shigenori Kanaya. Gene Cloning of an Alcohol Dehydrogenase from Thermophilic Alkane-Degrading Bacillus thermoleovorans B23. Journal of Bioscience and Bioengineering 91(1):100-102 (2001)
[4] Tomohisa Kato, Asuka Miyanaga, Shigenori Kanaya, Masaaki Morikawa. Gene cloning and characterization of an aldehyde dehydrogenase from long-chain alkane-degrading Geobacillus thermoleovorans B23. Extremophiles 14:33-39 (2010)
 "
"