Team:Newcastle
From 2010.igem.org
Swoodhouse (Talk | contribs) (→Project Description) |
|||
| (80 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__{{Team:Newcastle/mainbanner}} | __NOTOC__{{Team:Newcastle/mainbanner}} | ||
| - | =BacillaFilla: Fixing | + | =BacillaFilla: Fixing Cracks in Concrete= |
| - | + | ||
| - | + | ||
| + | [[Image:Newcastle_iGEM_Teampic.jpeg|centre|468px]] | ||
<div style="text-align:justify;padding:5px"> | <div style="text-align:justify;padding:5px"> | ||
===Project Description=== | ===Project Description=== | ||
| - | |||
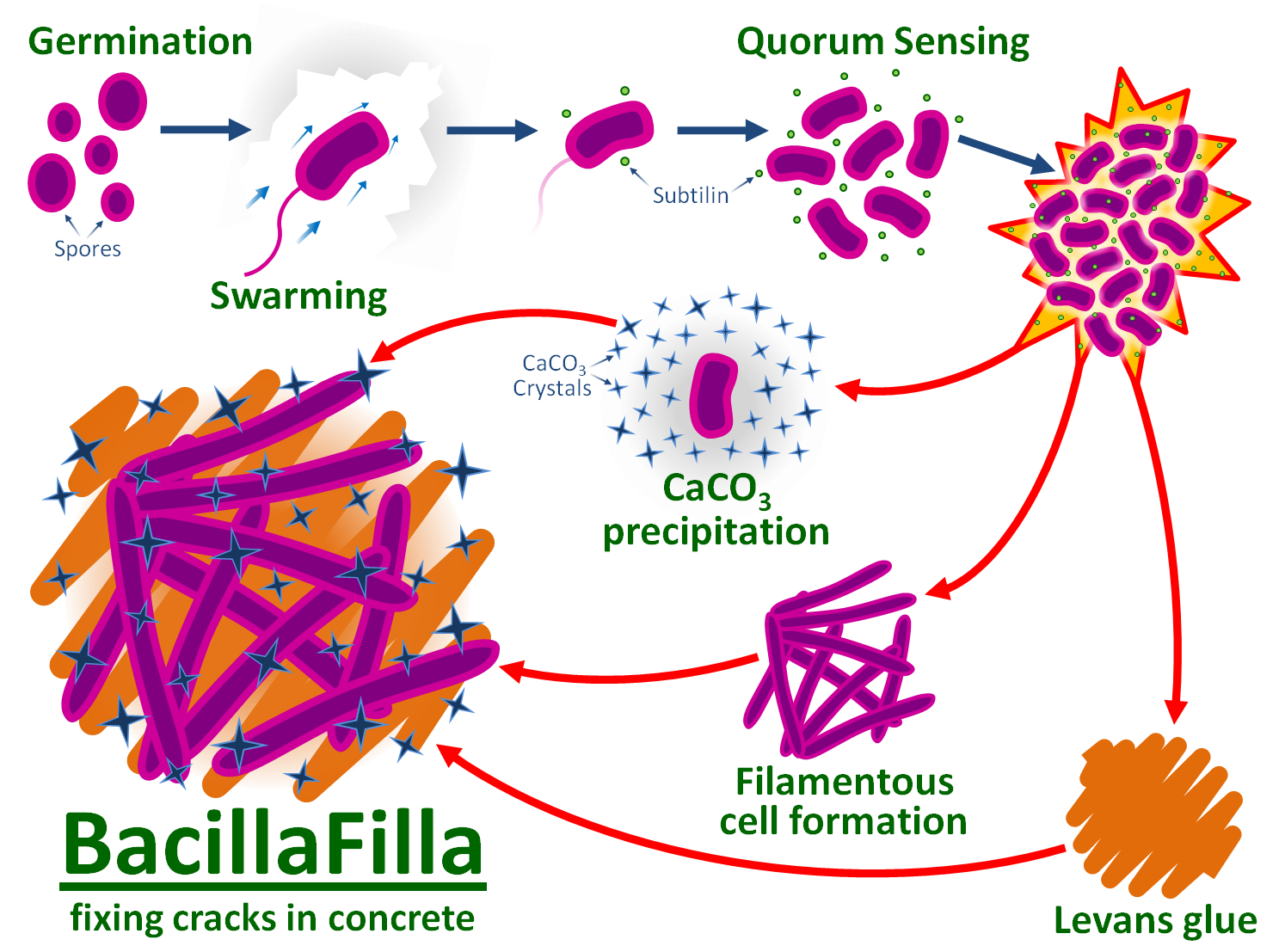
| - | BacillaFilla | + | BacillaFilla, an engineered ''Bacillus subtilis'', aims to repair [[Team:Newcastle/problem|cracks in concrete]] which can cause catastrophic structural failure. BacillaFilla would be applied to structures by spraying onto their surfaces. |
| - | ''B. subtilis'' 168 sporulates, making it ideal for storage and transportation. The cells | + | |
| + | BacillaFilla would swim deep into the cracks. Repair would be effected by production of [[Team:Newcastle/Urease|CaCO<sub>3</sub>]], [[Team:Newcastle/Filamentous_Cells|filamentous ''B. subtilis'' cells]] and [[Team:Newcastle/glue|levansucrose glue]]. CaCO<sub>3</sub> expands at the same rate as concrete, making it an ideal filler. Filamentous ''B. subtilis'' cells have similar tensile strength to the synthetic fibres used in fibre-reinforced concrete, and provide reinforcement. Levansucrose glues CaCO<sub>3</sub> and filamentous cells in place. | ||
| + | |||
| + | ''B. subtilis'' 168 sporulates, making it ideal for storage and transportation. The cells can be [[Team:Newcastle/solution#Alkalinity_resistance|made to be tolerant to concrete's high pH]]. | ||
| + | |||
| + | We designed a [[Team:Newcastle/Swarming|swarming]] BioBrick part for repairing ''B. subtilis'' 168's defective ''swrA'' and ''sfp'' genes, regaining motility. At the end of the crack the quorum sensing peptide [[Team:Newcastle/End_of_crack_%26_signalling_system|subtilin]] triggers a co-ordinated population response from a [[Team:Newcastle/End_of_crack_%26_signalling_system#2008Brick|subtilin-inducible promoter]]. Upregulating ''SR1'' and ''rocF'' promotes arginine and urea production, increasing exogenous CaCO<sub>3</sub> deposition. Over-producing YneA induces the filamentous cell phenotype, while SacB converts extracellular sucrose to levan glue. | ||
| + | |||
| + | To protect the environment our project also includes a design for a [[Team:Newcastle/Non-target-environment_kill_switch| kill switch]]. | ||
| + | |||
| + | Our project is summarised in the diagram below: | ||
| + | |||
| + | [[Image:newcastle_summary.png|600px|centre]] | ||
| + | |||
| - | |||
| + | ===Summary of Our Achievements=== | ||
| - | {|style cellpadding=" | + | {|style cellpadding="14" cellspacing="0" |
| - | + | ||
|- | |- | ||
| - | |<span style="color:Sienna">''' | + | |<span style="color:Sienna">'''BRONZE:'''</span> |
| - | | | + | |From the beginning of the project, we have successfully registered the team of two instructors, six advisors and eight members. We have completed and submitted a Project Summary form, developed ideas and shared them on our iGEM wikipedia page. We also entered 31 new BioBrick parts into the Registry of Parts. One part, [http://partsregistry.org/Part:BBa_K302012 IPTG-induced filamentous cell formation], the BioBrick for the IPTG-induced filamentous cell formation, was demonstrated to work as expected. |
|- | |- | ||
| - | |<span style="color:Silver">''' | + | |<span style="color:Silver">'''SILVER:'''</span> |
| - | | | + | |Our new BioBrick, the IPTG-inducible filamentous cell formation part works, so we [[Team:Newcastle/Filamentous_Cells#Characterisation|characterised]] it and included the information, [http://partsregistry.org/Part:BBa_K302012:Experience BBa_K302012], on the Parts Registry. |
|- | |- | ||
| - | |<span style="color:Goldenrod">''' | + | |<span style="color:Goldenrod">'''GOLD:'''</span> |
| - | | | + | |In order to obtain a gold, we investigated the benefits of an e-Science approach, focusing on workflows, to synthetic biology. Details can be found at [[Team:Newcastle/E-Science|here]]. This part of the project resulted in us proposing a new standard for a RESTful API which facilitates the discovery and publication of models of functional biological units. The standard has been submitted to the BioBricks Foundation as [[BBFRFC66|BBF RFC 66]]. |
|- | |- | ||
|} | |} | ||
Latest revision as of 19:13, 25 November 2010

| |||||||||||||
| |||||||||||||
BacillaFilla: Fixing Cracks in Concrete
Project Description
BacillaFilla, an engineered Bacillus subtilis, aims to repair cracks in concrete which can cause catastrophic structural failure. BacillaFilla would be applied to structures by spraying onto their surfaces.
BacillaFilla would swim deep into the cracks. Repair would be effected by production of CaCO3, filamentous B. subtilis cells and levansucrose glue. CaCO3 expands at the same rate as concrete, making it an ideal filler. Filamentous B. subtilis cells have similar tensile strength to the synthetic fibres used in fibre-reinforced concrete, and provide reinforcement. Levansucrose glues CaCO3 and filamentous cells in place.
B. subtilis 168 sporulates, making it ideal for storage and transportation. The cells can be made to be tolerant to concrete's high pH.
We designed a swarming BioBrick part for repairing B. subtilis 168's defective swrA and sfp genes, regaining motility. At the end of the crack the quorum sensing peptide subtilin triggers a co-ordinated population response from a subtilin-inducible promoter. Upregulating SR1 and rocF promotes arginine and urea production, increasing exogenous CaCO3 deposition. Over-producing YneA induces the filamentous cell phenotype, while SacB converts extracellular sucrose to levan glue.
To protect the environment our project also includes a design for a kill switch.
Our project is summarised in the diagram below:
Summary of Our Achievements
| BRONZE: | From the beginning of the project, we have successfully registered the team of two instructors, six advisors and eight members. We have completed and submitted a Project Summary form, developed ideas and shared them on our iGEM wikipedia page. We also entered 31 new BioBrick parts into the Registry of Parts. One part, [http://partsregistry.org/Part:BBa_K302012 IPTG-induced filamentous cell formation], the BioBrick for the IPTG-induced filamentous cell formation, was demonstrated to work as expected. |
| SILVER: | Our new BioBrick, the IPTG-inducible filamentous cell formation part works, so we characterised it and included the information, [http://partsregistry.org/Part:BBa_K302012:Experience BBa_K302012], on the Parts Registry. |
| GOLD: | In order to obtain a gold, we investigated the benefits of an e-Science approach, focusing on workflows, to synthetic biology. Details can be found at here. This part of the project resulted in us proposing a new standard for a RESTful API which facilitates the discovery and publication of models of functional biological units. The standard has been submitted to the BioBricks Foundation as BBF RFC 66. |
 
|
 "
"