Team:Imperial College London/Modules/Fast Response
From 2010.igem.org
(→Catechol-2,3-dioxygenase) |
(→Catechol-2,3-dioxygenase) |
||
| Line 29: | Line 29: | ||
TEV protease would then act upon its pool of inactive substrate already present in the cell. This substrate was an inactive form of the chose output enzyme Catechol 2,3 dioxygenase. | TEV protease would then act upon its pool of inactive substrate already present in the cell. This substrate was an inactive form of the chose output enzyme Catechol 2,3 dioxygenase. | ||
| - | ==Catechol-2,3-dioxygenase== | + | ==Catechol-2,3-dioxygenase enzyme and GFP-C(2,3)O fusion protein== |
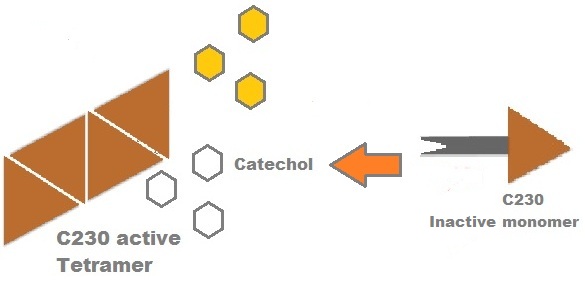
Catechol-2,3-dioxygenase protein is the product of XylE gene. It originates from Pseudomonas ''putida'' bacterium and the active protein is made of a homotetramer of monomers. The enzyme catlyzes the break down of catechol ring structure to produce 2-hydroxymuconic semialdehyde, a yellow colored product. | Catechol-2,3-dioxygenase protein is the product of XylE gene. It originates from Pseudomonas ''putida'' bacterium and the active protein is made of a homotetramer of monomers. The enzyme catlyzes the break down of catechol ring structure to produce 2-hydroxymuconic semialdehyde, a yellow colored product. | ||
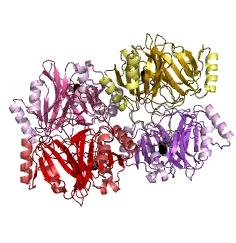
[[Image:Tetramer monomer C23O.jpg|thumb|300px|center|This figure illustrates the point that Catechol-2,3-dioxygenase enzyme is active only when it tetramerises]] | [[Image:Tetramer monomer C23O.jpg|thumb|300px|center|This figure illustrates the point that Catechol-2,3-dioxygenase enzyme is active only when it tetramerises]] | ||
Revision as of 21:58, 21 October 2010
| Modules | Overview | Detection | Signaling | Fast Response |
| Our design consists of three modules; Detection, Signaling and a Fast Response, each of which can be exchanged with other systems. We used a combination of modelling and human practices to define our specifications. Take a look at the overview page to get a feel for the outline, then head to the full module pages to find out how we did it. | |
| Fast Response Module |
|
Overview of the output module |
| TEV |
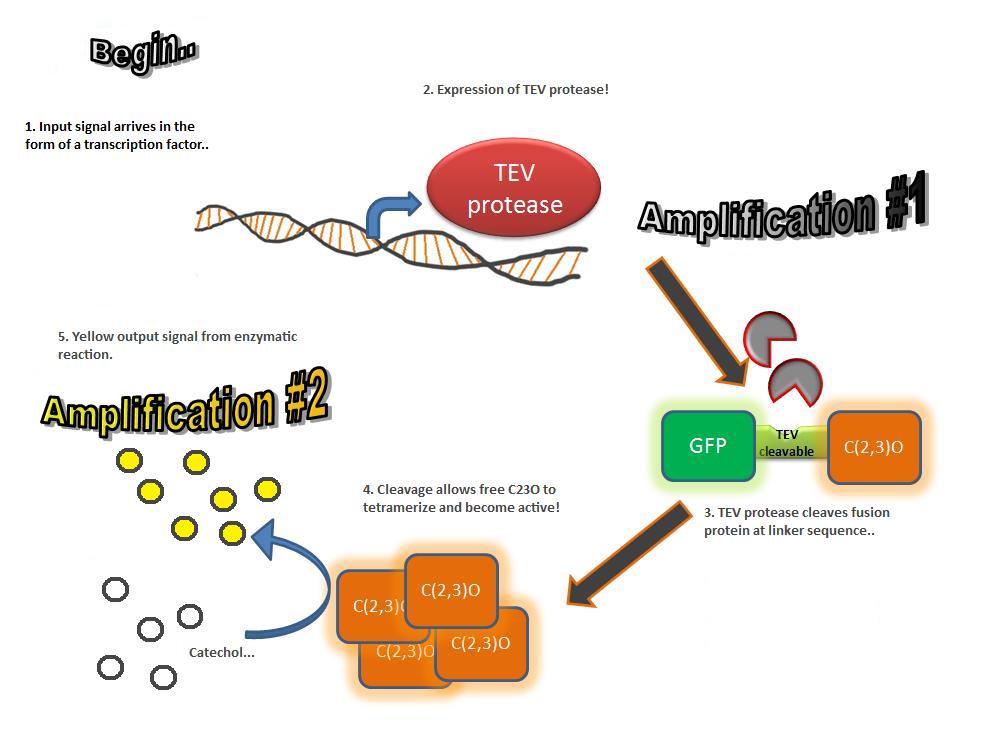
TEV proteaseTEV protease is a natural polyprotein cutting viral protease. In our system ComE transcription factors bind to their native ComCDE promoter which was engineered as the promoter for TEV protease. This means that TEV protease is only transcribed and translated upon detection of the Schistosoma parasite. Why TEV protease? This enzyme has been used extensively in protein engineering as it has a number of key qualities including a high specificity (ENYLFQG), and importantly for our system it is relatively tiny (242 aa) compared to other protease candidates available. Below is can be visualized as it has been deposited in the PDB with accession number 1Q31;
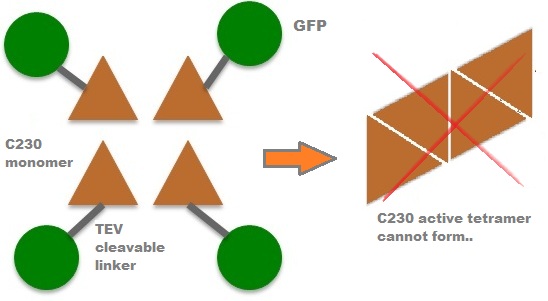
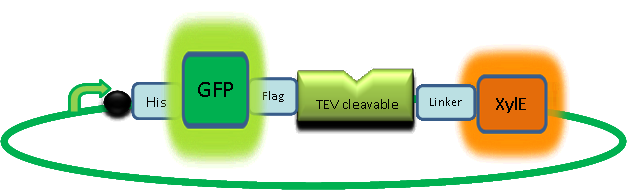
Due to its small size it’s likely that transcription and translation of the gene will not introduce limiting kinetic factors within our fast response system. TEV protease would then act upon its pool of inactive substrate already present in the cell. This substrate was an inactive form of the chose output enzyme Catechol 2,3 dioxygenase. Catechol-2,3-dioxygenase enzyme and GFP-C(2,3)O fusion proteinCatechol-2,3-dioxygenase protein is the product of XylE gene. It originates from Pseudomonas putida bacterium and the active protein is made of a homotetramer of monomers. The enzyme catlyzes the break down of catechol ring structure to produce 2-hydroxymuconic semialdehyde, a yellow colored product. In our project, we take advantage of the fact that Catechol-2,3-dioxygenase is only active as a tetramer. We have constructed a gene product that encodes a fussion protein. This fusion protein is made of GFP fused to the N-terminal of Catechol-2,3-dioxygenase. The GFP does not allow Catechol-2,3-dioxygenase monomers to associate by steric hindrance, so that the active tetramer is not formed. Instead the fusion protein remain as monomers in the cytosol of the cell. A very important feature of the fusion protein is its potential for induction of the system. A closer look at the fusion gene construct reveals the secret of the inducibility of our system. The GFP gene is fused to the XylE gene through a protein sequence that is susceptible to TEV protease. This protein sequence is recognized by the active site of the TEV protease so that when TEV is present in the cell, GFP is cleaved off Catechol-2,3-dioxygenase monomers. The free monomers are not longer stericly hindered by the GFP and are now able to associate with each other. Tetramerization of the monomers produces the active Catechol-2,3-dioxygenase enzyme, which in the presence of catechol substrate produces a yellow color. C23O is a homotetramer and its crystal structure has been solved. See Kita et al here it is viewed with the accession code 1MPY. For our fast response module we have taken advantage of the tetramerization feature which produces the functioning C230 enzyme; the active site forms as a result of intersubunit and interface interactions after oligomerization; We have made a fusion protein by adding GFP to the N- terminus, with the two connected by a linker cleavable by TEV protease. We hoped that this modified monomer would be unable to undergo oligomerisation until the TEV protease is activated. TEV would specifically cleave the linker between GFP and C23O, resulting in C23O monomers that are capable of oligomerising. Homotetramers would then be active and could catalyse the chromogenic reaction. The N-terminus was favoured over the C-terminus for modification because it is more accessible to the protease with it being positioned on the external surface of the monomer. However, it is further away from the oligomerisation interfaces and so might not actually prevent oligomerisation. The C-terminus, on the other hand, is less accessible, and by fusing GFP there it could prevent entry of the substrate into the funnel that runs through the enzyme. It is closer to the oligomerisation domain and so fusing GFP would be more likely to prevent oligomerisation. After observing C23O in Pymol, and in addition information obtained from Kita et al which shows that the projecting loop is needed for dimerization and that this is very near to the N-terminus, we chose to engineer (by straightforward DNA synthesis) GFP plus linker (GGGSGGGS ENYLFQG) onto the N-termini of our C230 monomers.
Image taken from ftp://ftp.genome.jp/pub/kegg Where catechol is the colourless substrate converted by ring cleavage into 2 hydroxymuconate semialdehyde. |
| C23O |
Catechol 2,3 dioxygenaseCatechol 2,3-dioxygenase (also known as C23O or MPC) catalyses the conversion of a colourless substrate (catechol or substituted catechols) into a bright yellow product (2-hydroxymuconic semialdehyde) within seconds of substrate addition. We performed our specific plate assays using a 100mM stock solution of catechol and via Pasteur pipette covering individual colonies in the solution. Liquid assays were performed with addition of a pre-made known concentration of catechol typically 0.05-0.35M diluted with DDH20. It was convenient that a Biobrick for the XylE gene was already present in the registry and we used this along with biobrick cloning methods to generate many of our XylE final and testing constructs. An issue with our chosen enzyme C23O was that we weren’t sure whether it could be successfully expressed in B.subtilis as most papers referred to assays within an E.coli host. However it was later found to have been expressed successfully in B. subtilis by Zukowski et al. |
|
Here's a picture of the final construct: |
References
[1]An archetypical extradiol-cleaving catecholic dioxygenase: the crystal structure of catechol 2,3-dioxygenase (metapyrocatechase) from Pseudomonas putida mt-2 Akiko Kita1, Shin-ichi Kita2, Ikuhide Fujisawa1, Koji Inaka3, Tetsuo Ishida4, Kihachiro Horiike4, Mitsuhiro Nozaki4 and Kunio Miki5,
 "
"