Team:Imperial College London/Parts/Favourites
From 2010.igem.org
(New page: {{:Team:Imperial_College_London/Templates/Header}}) |
|||
| (8 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Imperial_College_London/Templates/Header}} | {{:Team:Imperial_College_London/Templates/Header}} | ||
| + | {{:Team:Imperial_College_London/Templates/PartsHeader}} | ||
| + | |||
| + | {| style="width:900px;background:#f5f5f5;text-align:left;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|XylE - catechol 2,3-dioxygenase from P.putida with terminator | ||
| + | |- | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|[http://partsregistry.org/Part:BBa_K316003 '''BBa_K316003'''] | ||
| + | |- | ||
| + | |Catechol or catechol 2,3-dioxygenases (C2,3O) + O(2) is converted by a ring cleavage into 2-hydroxymuconate semialdehyde which is the cytotoxic and bright yellow-coloured product<cite>1</cite>. This is a key enzyme in many (soil) bacterial species used for the degradation of aromatic compounds. Catechol 2,3-dioxygenase<cite>2</cite> was originally isolated from Pseudomonas putida and is active only as a homotetramer. Tetramerization, mediated via beta-sheets in the N-terminal regions of the monomers, allows formation of the active site which covalently binds a ferrous ion. The reaction itself takes place within seconds after the addition by Pasteur pipette or spraying of catechol at a 100mM stock solution diluted with DDH20 (used by our lab.) The toxic breakdown product is thought to interfere with cell wall integrity and cellular machinery such that exposed cells gradually die. | ||
| + | |||
| + | |||
| + | '''Safety''' | ||
| + | |||
| + | Catechol is classed as irritant in the EU but as toxic in the USA, as well as being a possible carcinogen. It should therefore be handled with care and proper safety equipment. More information is available on the Material Safety Data Sheet[http://www.sciencelab.com/msds.php?msdsId=9927131]. | ||
| + | |||
| + | |||
| + | |||
| + | <span class='h3bb'><big>'''Sequence and Features'''</big></span> | ||
| + | |||
| + | <partinfo>BBa_K316003 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <span class='h3bb'><big>'''Part Characterisation'''</big></span> | ||
| + | |||
| + | {| | ||
| + | |colspan="2"| | ||
| + | |- | ||
| + | | | ||
| + | <span class='h3bb'><big>'''Optimum absorbtion wavelength for catechol assays'''</big></span> | ||
| + | |||
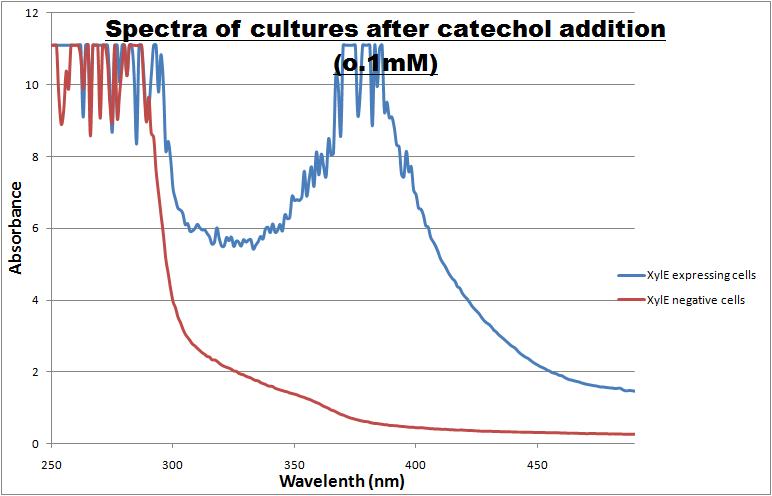
| + | [[Image:Spectra of Xyle cells.jpg|thumb|right|400px| Peak absorbance of catechol breakdown product (2-hydroxymuconic semialdehyde]] | ||
| + | |||
| + | The enzymatic reaction catalysed by XylE can also serve as a powerful reporter. The substrate - catechol - is colourless. However within seconds of its addition, the colonies or suspended cultures of XylE-expressing cells become clearly yellow<cite>3</cite> to the naked eye. This reaction allows direct measurement of XylE activity by measuring product concentrations, which absorbs light in the visible spectrum, at 380nm. In contrast to other common reporters like GFP, LacZ or Luciferase which do easily express correctly in thermophile environments, a XylE assay has also been shown to be functional in here. The spectrophotometric assay compared the spectra of two cultures of E.coli (one XylE gene transformed and the other not) were compared on addition of 0.1mM Catechol substrate. | ||
| + | |||
| + | A spectrophotometric assay of two cultures of E.coli (Blue: contains <bbpart>BBa_K316004</bbpart>, Red: not expressing XylE ) were compared on addition of 0.1mM Catechol substrate. The spectra show that in XylE transformed cells, a broad peak appears at about 380nm. The absorbance at this particular wavelength is due to the yellow product of the reaction (2-hydroxymuconic semialdehyde (HMS)). | ||
| + | | | ||
| + | |} | ||
| + | {| | ||
| + | |colspan="2"| | ||
| + | |- | ||
| + | | | ||
| + | <span class='h3bb'><big>'''''In Vitro'' Assay'''</big></span> | ||
| + | |||
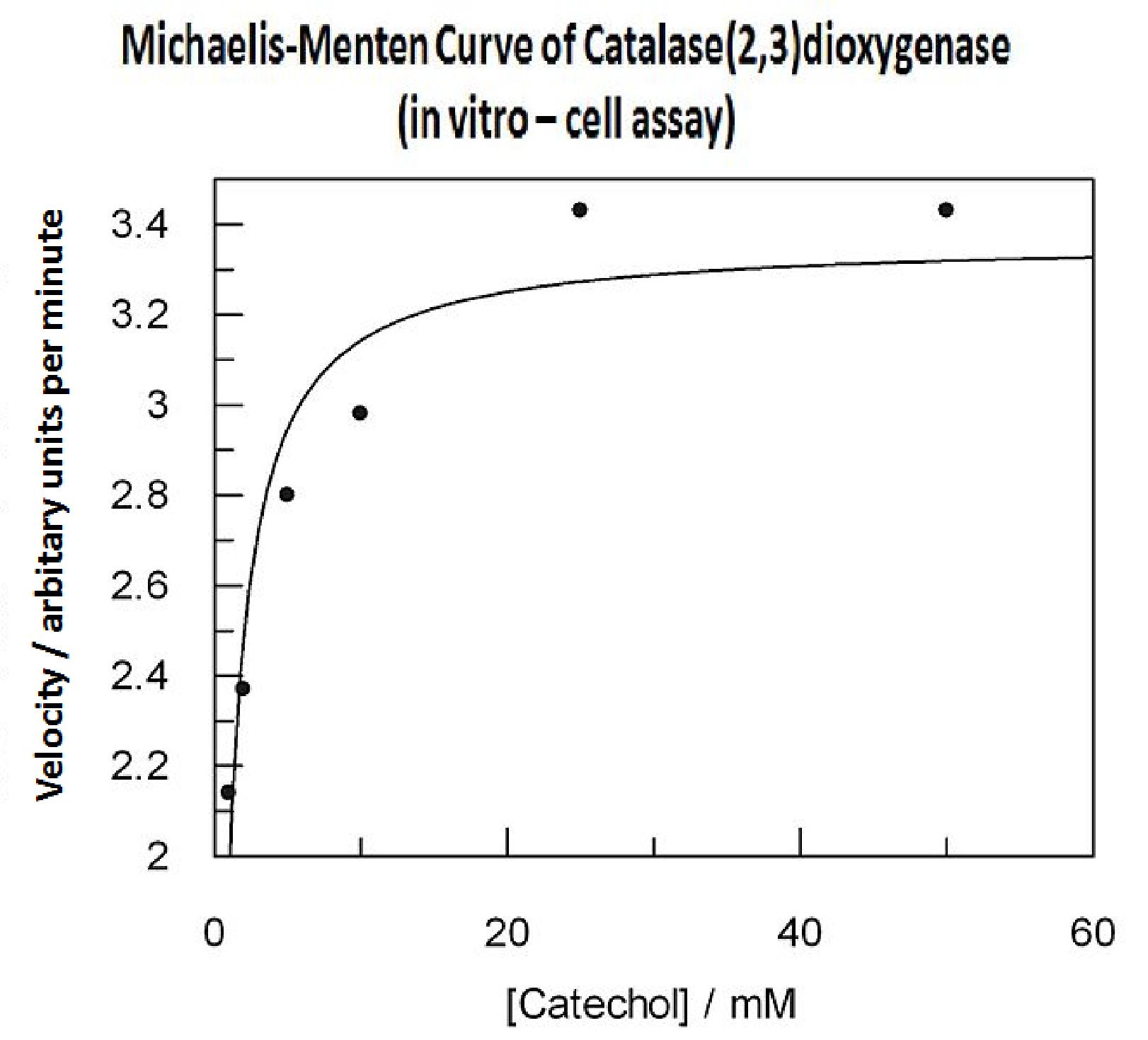
| + | [[Image:Grapfit curve 1.jpg|thumb|right|400px]] | ||
| + | |||
| + | Due to technical limitations, to measure kinetic parameters of XylE is to lyse cells and . In this experiment cell lysate was assayed with increasing catechol concentrations. The rate at which the yellow product appears is directly proportional to the velocity of the reaction. The rate reaction was monitored by measuring color output of the reaction in the plate reader. | ||
| + | |||
| + | Cell lysate was tested for dioxygenase activity to determine appropriate dilutions for the assay. The cell lysate was obtained from a 100ml overnight culture and diluted by a factor of 20 to obtain a suitable concentration of total enzyme for the plate reader assay. The concentrations of catechol used were 1, 2, 5, 10, 25, 50 mM. | ||
| + | |||
| + | Data collected was used to construct the Michaelis-Menten curve for the in vitro kinetics of XylE in cell lysate. | ||
| + | Michaelis-Menten curve was drawn using velocity values calculated from the slope at the initial stages of the reaction, as this is the only time when substrate concentration values are accurate. The plot was delineated by non-linear regression analysis using GraFit software tool[http://www.erithacus.com/grafit/]. The calculated Km is 0.71mM catechol (with a Vmax of 3.37 in O.D. arbitrary units for this dilution of cell lysate). | ||
| + | |||
| + | For more detailed information, please check our Results section [[Team:Imperial_College_London/Results/Exp6]] | ||
| + | | | ||
| + | |} | ||
| + | {| | ||
| + | |colspan="2"| | ||
| + | |- | ||
| + | | | ||
| + | <span class='h3bb'><big>'''Cytotoxicity of Catechol'''</big></span> | ||
| + | |||
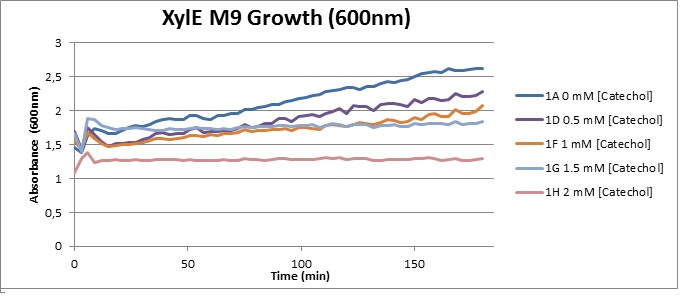
| + | [[Image:XylE M9 Growth (600).jpg|thumb|right|400px|O.D. at 600 over 3h for XylE-transformed Top10 cells in presence of different catechol concentrations, growing in M9 medium.]] | ||
| + | |||
| + | The addition of catechol had distinctive deleterious effects on XylE expressing cells. While at 0% catechol growth-behavior did not show a significant change (dark blue), even the lowest concentration of 0.25% catechol appeared to drastically reduce cell-survival (red). In contrast, CMR-control cells did not change their growing behavior in the presence of catechol. From this we conclude that the breakdown product of catechol, 2-hydroxymuconic semialdehyde, and not Catechol itself, has strong cytotoxic effects | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | ---- | ||
| + | Characterisation data was obtained using GFP-XylE constructs <bbpart>BBa_K316007</bbpart> and XylE under two different promoters: ''B. subtilis'' derived Pveg <bbpart>BBa_K316005</bbpart> and J23101 <bbpart>BBa_K316004</bbpart> from ''E. coli''. These are described on our wiki[https://2010.igem.org/Team:Imperial_College_London/Results] and the aforementioned parts pages. | ||
| + | |||
| + | |||
| + | ===References=== | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/10368270 Kita ''et al.'' 1999] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/12519074 Okuta ''et al.'' 2003] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/6405380 Zukowski ''et al.'' 1983] | ||
| + | |} | ||
| + | {| style="width:900px;background:#f5f5f5;text-align:left;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|B. subtilis transformation vector with LacI, targets amyE locus | ||
| + | |- | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|[http://partsregistry.org/Part:BBBa K316027 '''BBBa K316027'''] | ||
| + | |- | ||
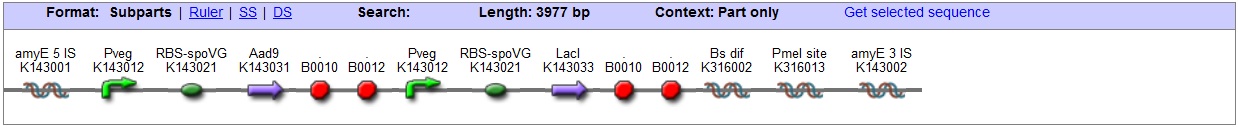
| + | |This vector has been designed using the amyE 5' and 3' integration sequences for integration into ''B.subtilis'' genome | ||
| + | |||
| + | |||
| + | '''AmyE locus''' | ||
| + | This vector has been designed using the amyE 5' <bbpart>BBa_K143008</bbpart> and 3 <bbpart>BBa_K143009</bbpart>' integration sequences for integration into ''B. subtilis'' genome. Insertion into the amyE locus provides a selection marker as the bacterium will no longer be able to breakdown starch. An iodine assay can be used to confirm integration. This phenotype makes the transformed bacterium considerably less likely to survive in natural environments. | ||
| + | |||
| + | |||
| + | '''LacI''' | ||
| + | With constitutivly expressed LacI <bbpart>BBa_K143033</bbpart> the vector is suitable for ''B. subtilis'' transformations with hyper-spank <bbpart>BBa_K143015</bbpart> controlled gene-circuits. | ||
| + | |||
| + | '''Chloramphenicol Resistance''' | ||
| + | This vector also contains a positive selection marker, flanked by two dif sites. Chloramphenicol acetyltransferase <bbpart>BBa_J31005</bbpart> provides resistance to chloramphenicol antibiotic. | ||
| + | |||
| + | '''Blunt end cloning site''' | ||
| + | PmeI restriction site <bbpart>BBa_K316013</bbpart> is designed as a cloning site. Due to the 8bp recognition sequence it is a rare site that can be used to cut the vector only once. | ||
| + | |||
| + | [[Image:AmyE LacI vector Graphic2.jpg|center|850px]] | ||
| + | |||
| + | |||
| + | [[Image:VectorLacI.PNG|center|800px]] | ||
| + | |||
| + | '''Figure I.''' Complete amyE vector for ''B. subtilis'' genome integration with LacI expression. | ||
| + | |||
| + | |} | ||
| + | {| style="width:900px;background:#f5f5f5;text-align:left;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Pveg-spoVG-LytC-Helical Linker-Elastase Cleavage Site-AIP-His Tag | ||
| + | |- | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|[http://partsregistry.org/Part:BBa_K316037 '''BBa_K316037'''] | ||
| + | |- | ||
| + | |'''Introduction:''' This part, our detection module, was designed to anchor a signal peptide (AIP) to the cell wall of B. subtilis which is released upon cleave of the part by a protease, in this case the cercarial elastase [http://www.uniprot.org/uniprot/P12546 '''(UniProt,'''] [http://merops.sanger.ac.uk/cgi-bin/pepsum?id=S01.144 '''MEROPS)'''], to activate our output. The part is fully assembled and ready to be expressed in a prokaryotic cell as it contains a promoter we characterised, an optimised RBS and a double stop codon. The protein product includes a cell wall binding domain (CWB) of LytC, [http://partsregistry.org/Part:BBa_K316030 BBa_K316030] a linker that has been designed to carry a protease cleave site and the AIP [http://partsregistry.org/Part:BBa_K316047 BBa_K316047]. This part is essential for the great modularity of our system, as a change of just 4 amino acids can be used to confer specificity to another protease see our [https://2010.igem.org/Team:Imperial_College_London/Software_Tool '''Software Tool''']. | ||
| + | |||
| + | |||
| + | '''Pveg-spoVG:''' This was build in 2008 by the Imperial College iGEM team [http://partsregistry.org/Part:BBa_K143053 BBa_K143053]. | ||
| + | |||
| + | '''LytC:''' The CWB is on the 5’ end and anchors the detection module to the cell wall of B. subtilis. The CWB was separately submitted as biobrick [http://partsregistry.org/Part:BBa_K316030 BBa_K316030]. | ||
| + | |||
| + | '''
Helical Linker:''' The Linker separates the CWB and the AIP and creates space for the protease to access the cleavage site; it consists of two main sections. The first six amino acids (SRGSRA) were suggested to be used specifically with LytC [http://www.ncbi.nlm.nih.gov/pubmed/14594841 Yamamoto ''et al.'' 2003]. The second section consists of a helical amino acid sequence that is stiff and prevents interaction of the AIP with its receptor and thus false positive activation of the output system. It was specifically designed to separate protein domains [http://www.ncbi.nlm.nih.gov/pubmed/11579220 Arai ''et al.'' 2001]. | ||
| + | |||
| + | '''
Elastase Cleavage Site:''' This sequence forms the 3’ end of the linker and is directly attached to the 5’ end of the AIP. It is four amino acids (SWPL) long and was designed to be efficiently cleaved by the schistosoma cercarial elastase [http://www.jbc.org/content/277/27/24618.abstract Salter ''et al.'' 2002]. | ||
| + | |||
| + |
'''AIP:''' The linear auto-inducing peptide (AIP) from the Streptococcus pneumonia ComCDE system, called Competence-Stimulating Peptide-1 (CSP-1 or ComC) . Upon cleavage it is free to diffuse and bind it's receptor ComD [http://partsregistry.org/Part:BBa_K316015 BBa_K316015]. Signalling cascade from ComD is then able to activate transcription of genes under specific promoter sequences. | ||
| + | |||
| + |
'''His-Tag:''' To be able to purify the protein for testing, we attached a His-Tag on our linker-AIP peptide. As it would probably interfere with recognition of the AIP by the receptor it has to be removed from the final construct.
| ||
| + | |||
| + | '''Stop Codon:''' In order to end translation a double stop codon was put in place. | ||
| + | |||
| + | [[Image:Surface protein BB graphics.jpg|center|850px]] | ||
| + | |||
| + | '''Diagramatic representation of surface protein.''' | ||
| + | |||
| + | [[Image:PromoterCWBprot.png|center|600px]] | ||
Latest revision as of 01:21, 28 October 2010
| Parts | Favourites | Full List |
| We have contributed 23 parts to the Registry of Standard Biological Parts and we really hope that other people will find them useful in the future. Here you can find detailed information on our favourite parts, or the registry information on the complete parts list. | |
| XylE - catechol 2,3-dioxygenase from P.putida with terminator | ||||||||||||
| [http://partsregistry.org/Part:BBa_K316003 BBa_K316003] | ||||||||||||
| Catechol or catechol 2,3-dioxygenases (C2,3O) + O(2) is converted by a ring cleavage into 2-hydroxymuconate semialdehyde which is the cytotoxic and bright yellow-coloured product1. This is a key enzyme in many (soil) bacterial species used for the degradation of aromatic compounds. Catechol 2,3-dioxygenase2 was originally isolated from Pseudomonas putida and is active only as a homotetramer. Tetramerization, mediated via beta-sheets in the N-terminal regions of the monomers, allows formation of the active site which covalently binds a ferrous ion. The reaction itself takes place within seconds after the addition by Pasteur pipette or spraying of catechol at a 100mM stock solution diluted with DDH20 (used by our lab.) The toxic breakdown product is thought to interfere with cell wall integrity and cellular machinery such that exposed cells gradually die.
Catechol is classed as irritant in the EU but as toxic in the USA, as well as being a possible carcinogen. It should therefore be handled with care and proper safety equipment. More information is available on the Material Safety Data Sheet[http://www.sciencelab.com/msds.php?msdsId=9927131].
Sequence and Features <partinfo>BBa_K316003 SequenceAndFeatures</partinfo>
Characterisation data was obtained using GFP-XylE constructs <bbpart>BBa_K316007</bbpart> and XylE under two different promoters: B. subtilis derived Pveg <bbpart>BBa_K316005</bbpart> and J23101 <bbpart>BBa_K316004</bbpart> from E. coli. These are described on our wiki[1] and the aforementioned parts pages.
References[http://www.ncbi.nlm.nih.gov/pubmed/10368270 Kita et al. 1999] [http://www.ncbi.nlm.nih.gov/pubmed/12519074 Okuta et al. 2003] [http://www.ncbi.nlm.nih.gov/pubmed/6405380 Zukowski et al. 1983] | ||||||||||||
| B. subtilis transformation vector with LacI, targets amyE locus |
| [http://partsregistry.org/Part:BBBa K316027 BBBa K316027] |
| This vector has been designed using the amyE 5' and 3' integration sequences for integration into B.subtilis genome
Chloramphenicol Resistance This vector also contains a positive selection marker, flanked by two dif sites. Chloramphenicol acetyltransferase <bbpart>BBa_J31005</bbpart> provides resistance to chloramphenicol antibiotic. Blunt end cloning site PmeI restriction site <bbpart>BBa_K316013</bbpart> is designed as a cloning site. Due to the 8bp recognition sequence it is a rare site that can be used to cut the vector only once.
Figure I. Complete amyE vector for B. subtilis genome integration with LacI expression. |
| Pveg-spoVG-LytC-Helical Linker-Elastase Cleavage Site-AIP-His Tag |
| [http://partsregistry.org/Part:BBa_K316037 BBa_K316037] |
| Introduction: This part, our detection module, was designed to anchor a signal peptide (AIP) to the cell wall of B. subtilis which is released upon cleave of the part by a protease, in this case the cercarial elastase [http://www.uniprot.org/uniprot/P12546 (UniProt,] [http://merops.sanger.ac.uk/cgi-bin/pepsum?id=S01.144 MEROPS)], to activate our output. The part is fully assembled and ready to be expressed in a prokaryotic cell as it contains a promoter we characterised, an optimised RBS and a double stop codon. The protein product includes a cell wall binding domain (CWB) of LytC, [http://partsregistry.org/Part:BBa_K316030 BBa_K316030] a linker that has been designed to carry a protease cleave site and the AIP [http://partsregistry.org/Part:BBa_K316047 BBa_K316047]. This part is essential for the great modularity of our system, as a change of just 4 amino acids can be used to confer specificity to another protease see our Software Tool.
Pveg-spoVG: This was build in 2008 by the Imperial College iGEM team [http://partsregistry.org/Part:BBa_K143053 BBa_K143053]. LytC: The CWB is on the 5’ end and anchors the detection module to the cell wall of B. subtilis. The CWB was separately submitted as biobrick [http://partsregistry.org/Part:BBa_K316030 BBa_K316030]. Helical Linker: The Linker separates the CWB and the AIP and creates space for the protease to access the cleavage site; it consists of two main sections. The first six amino acids (SRGSRA) were suggested to be used specifically with LytC [http://www.ncbi.nlm.nih.gov/pubmed/14594841 Yamamoto et al. 2003]. The second section consists of a helical amino acid sequence that is stiff and prevents interaction of the AIP with its receptor and thus false positive activation of the output system. It was specifically designed to separate protein domains [http://www.ncbi.nlm.nih.gov/pubmed/11579220 Arai et al. 2001]. Elastase Cleavage Site: This sequence forms the 3’ end of the linker and is directly attached to the 5’ end of the AIP. It is four amino acids (SWPL) long and was designed to be efficiently cleaved by the schistosoma cercarial elastase [http://www.jbc.org/content/277/27/24618.abstract Salter et al. 2002]. AIP: The linear auto-inducing peptide (AIP) from the Streptococcus pneumonia ComCDE system, called Competence-Stimulating Peptide-1 (CSP-1 or ComC) . Upon cleavage it is free to diffuse and bind it's receptor ComD [http://partsregistry.org/Part:BBa_K316015 BBa_K316015]. Signalling cascade from ComD is then able to activate transcription of genes under specific promoter sequences. His-Tag: To be able to purify the protein for testing, we attached a His-Tag on our linker-AIP peptide. As it would probably interfere with recognition of the AIP by the receptor it has to be removed from the final construct. Stop Codon: In order to end translation a double stop codon was put in place. Diagramatic representation of surface protein. |
 "
"