Team:uOttawa/Project
From 2010.igem.org
Djedrysiak (Talk | contribs) (→Power of yeast mating) |
Djedrysiak (Talk | contribs) (→Project Overview) |
||
| Line 9: | Line 9: | ||
==Project Overview== | ==Project Overview== | ||
| - | Yeast is one of the earliest organisms used in biotechnology, even if we did not know that we were doing science at the time. Humans have been experimenting with yeast ever since our ancestors brewed their first beer. However, there is more to yeast than just making beer and bread; it also happens to be one of the simplest eukaryotic organisms. Due to its homology with mammalian organisms and its ease of genetic manipulation, yeast has become a model organism for the study of systems biology. Many key genes in mammalian organisms were first identified in yeast. For this reason, we believe it is important to develop tools that improve our ability to use yeast as a standard BioBrick chassis. | + | Yeast (''Saccharomyces cerevisiae'') is one of the earliest organisms used in biotechnology, even if we did not know that we were doing science at the time. Humans have been experimenting with yeast ever since our ancestors brewed their first beer. However, there is more to yeast than just making beer and bread; it also happens to be one of the simplest eukaryotic organisms. Due to its homology with mammalian organisms and its ease of genetic manipulation, yeast has become a model organism for the study of systems biology. Many key genes in mammalian organisms were first identified in yeast. For this reason, we believe it is important to develop tools that improve our ability to use yeast as a standard BioBrick chassis. |
In summary we have BioBricked two yeast selection markers and have shown that they work, constructed a novel plasmid which greatly increases the efficiency of genomic integration into yeast, BioBricked several new yeast promoters and characterized one, developed a novel strategy for dissecting promoters. Please, browse through the following sections in order to learn more about each aspect of our project. We hope that these contributions will encourage more iGem teams to use yeast as part of their projects! | In summary we have BioBricked two yeast selection markers and have shown that they work, constructed a novel plasmid which greatly increases the efficiency of genomic integration into yeast, BioBricked several new yeast promoters and characterized one, developed a novel strategy for dissecting promoters. Please, browse through the following sections in order to learn more about each aspect of our project. We hope that these contributions will encourage more iGem teams to use yeast as part of their projects! | ||
Revision as of 03:30, 28 October 2010
 |
||||||||
Contents |
Project Overview
Yeast (Saccharomyces cerevisiae) is one of the earliest organisms used in biotechnology, even if we did not know that we were doing science at the time. Humans have been experimenting with yeast ever since our ancestors brewed their first beer. However, there is more to yeast than just making beer and bread; it also happens to be one of the simplest eukaryotic organisms. Due to its homology with mammalian organisms and its ease of genetic manipulation, yeast has become a model organism for the study of systems biology. Many key genes in mammalian organisms were first identified in yeast. For this reason, we believe it is important to develop tools that improve our ability to use yeast as a standard BioBrick chassis.
In summary we have BioBricked two yeast selection markers and have shown that they work, constructed a novel plasmid which greatly increases the efficiency of genomic integration into yeast, BioBricked several new yeast promoters and characterized one, developed a novel strategy for dissecting promoters. Please, browse through the following sections in order to learn more about each aspect of our project. We hope that these contributions will encourage more iGem teams to use yeast as part of their projects!
New selection markers
There are generally two types of selection markers used in yeast, auxotropic markers and selection markers that confer drug resistance. Standard yeast strains have deletions and mutations in genes in a pathway that generates an essential metabolite (such as an amino acid). These yeast strains must be grown on media supplemented with that particular metabolite. When integrating constructs into yeast, a functional copy of that gene can be brought over. Only yeast cells that contain that functional copy will be able to grow on media lacking that specific metabolite. From our experience we have learned that there are two major drawbacks with such selection. Firstly, this type of selection is not very strong, small background colonies that do not contain the integrated construct often appear on plates. Secondly, since the selection marker shares substantial homology with the mutated / partially deleted ORF in the wild type strain, it is possible that the selection marker can undergo homologous recombination with the wild type ORF and this results in a yeast colony that can survive on the restricted media but does not contain the construct of interest. Drug selection markers are much stronger than auxotrophic markers in yeast and for this reason they have less background on plates. We BioBricked natMX6 and kanMX, which are completely heterologous and do not share any homology with the yeast genome. There is less chance that they will recombinate into the incorrect locus. They provide resistance to the drugs Nourseothricin sulfate and G418 respectively.
Results
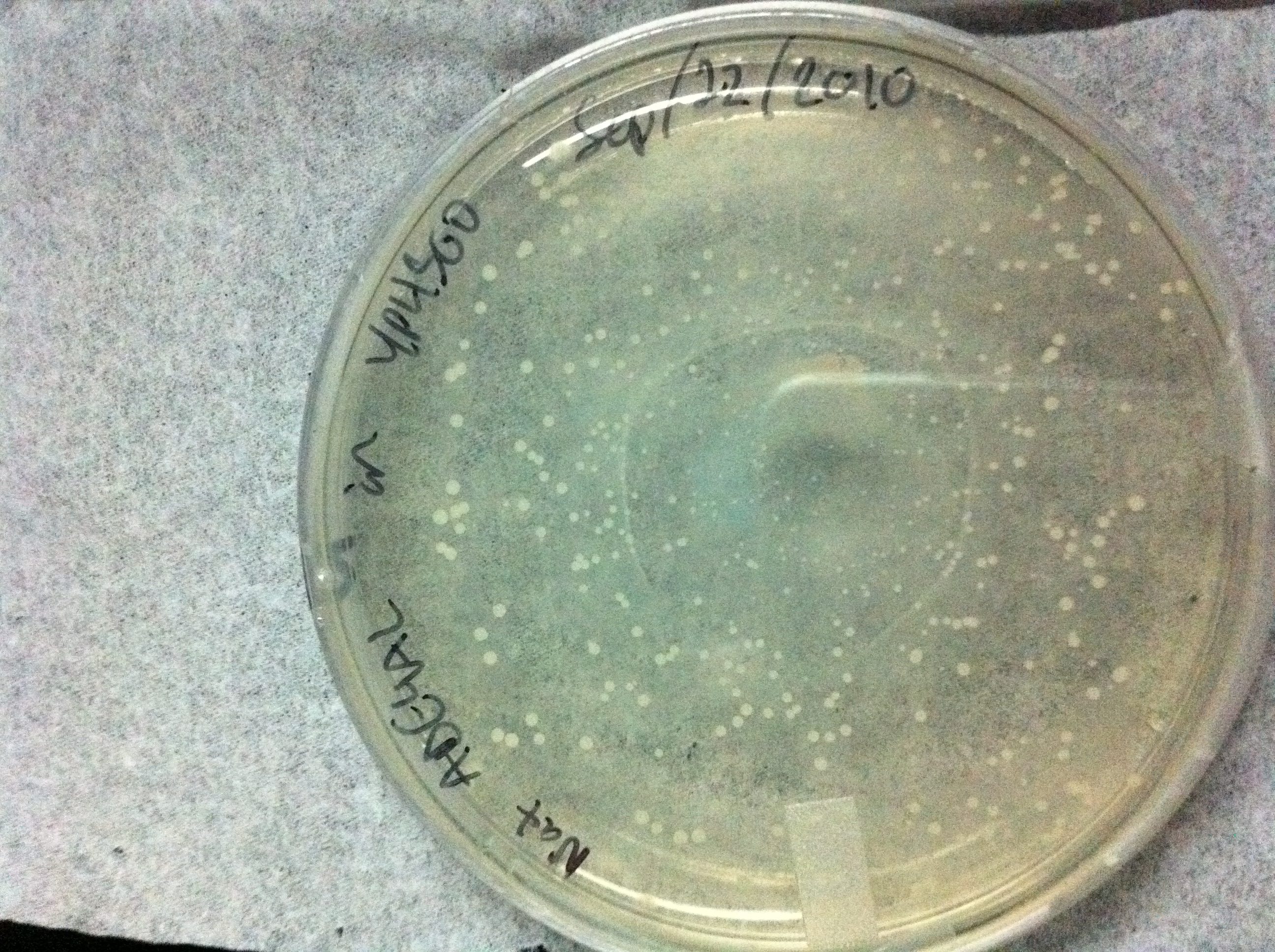
Figure 1:YPH500 cells transformed with our ADE4 targeting vector containing the natMX6 resistance marker. The plate contains YPD media with 100 ug/mL Nourseothricin. White colonies indicate a successful transformation into the ADE4 locus, all colonies are white.
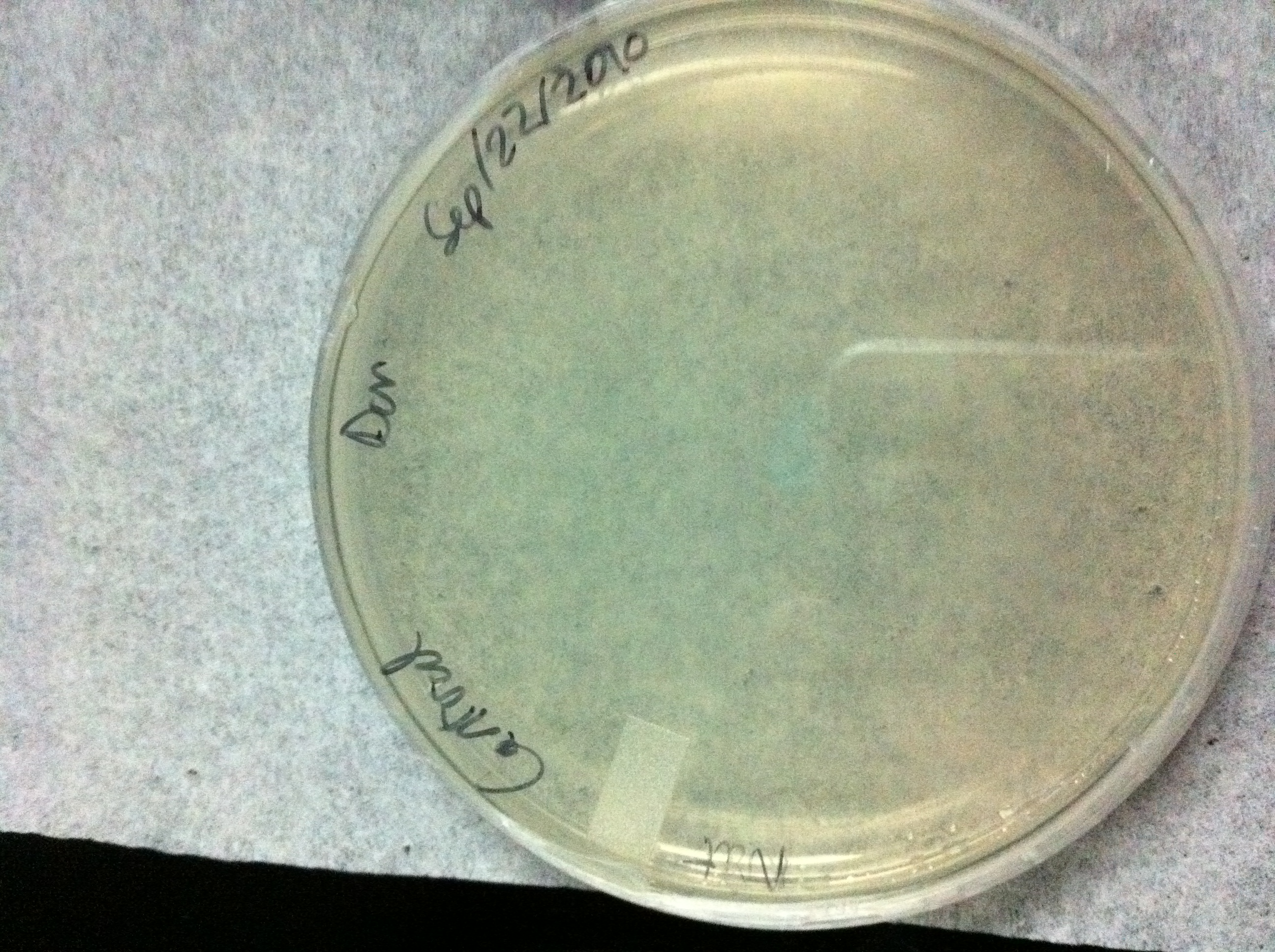
Figure 2:Wild type YPH500 cells plated on YPD media with 100 ug/mL Nourseothricin, no growth is observed
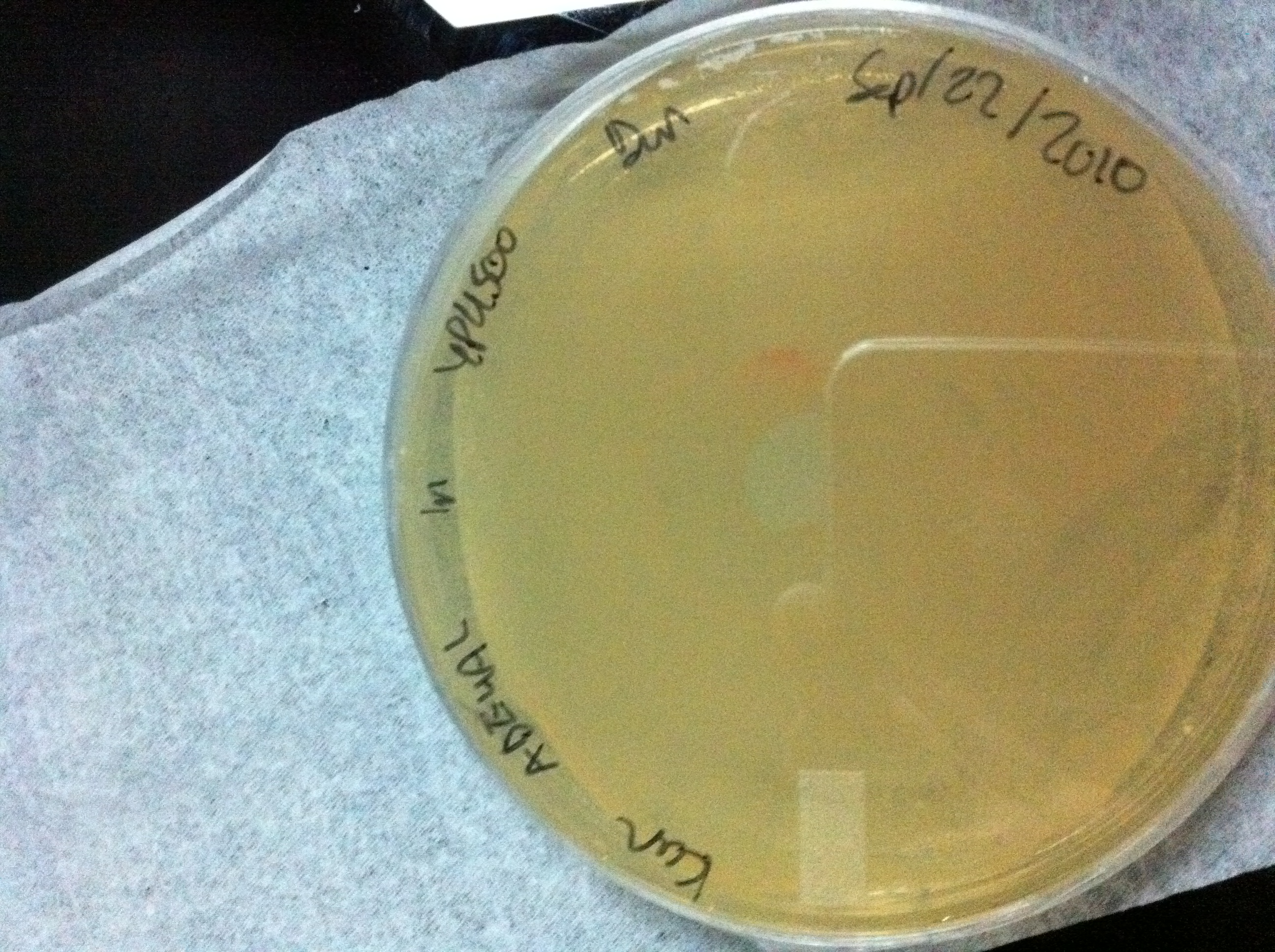
Figure 3:YPH500 cells transformed with our ADE4 targeting vector containing the kanMX resistance marker. The plate contains YPD media with 0.2 mg/mL G418. White colonies indicate a successful transformation into the ADE4 locus. Transformation efficiency is very high, single colonies are hard to discern, cells seem to be growing as a lawn. Areas of high cell density appear red presumably due to cells growing on top of each other. Areas of lower cell density are white.
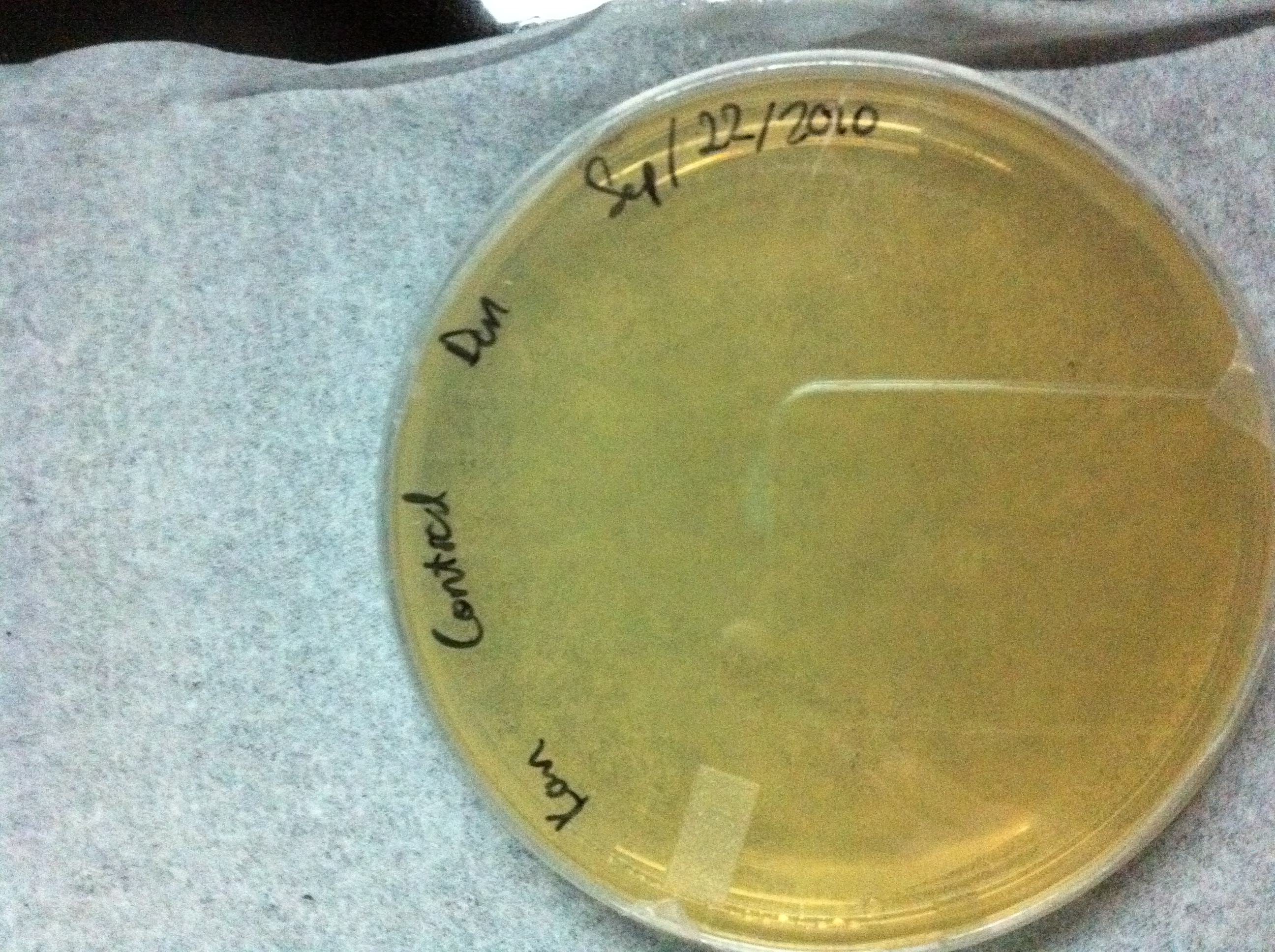
Figure 4:Wild type YPH500 cells plated on YPD media with 0.2 mg/mL , no growth is observed
Ade4 targeting vector
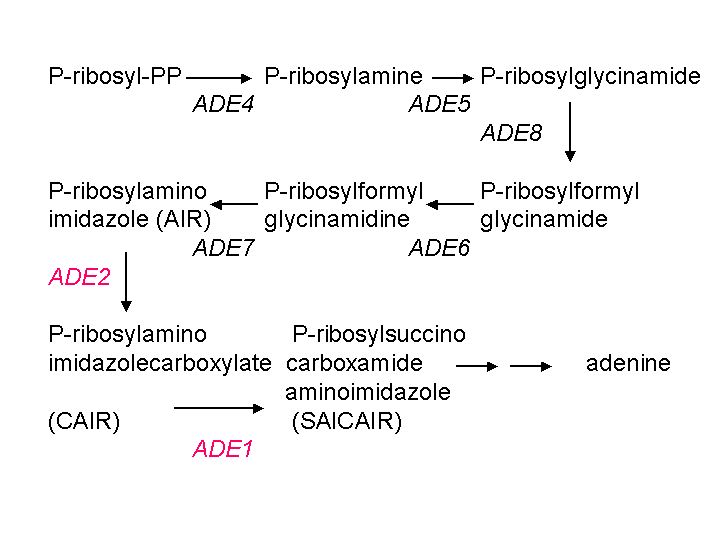
When implementing finely balanced synthetic networks in vivo it is very important to tightly control the copy number of network components (Guido, 2006). In yeast the easiest way to control the copy number is to keep it at 1 by integrating constructs into the yeast genome. Constructs can be integrated into the yeast genome by homologous recombination, which is a natural yeast mechanism used to repair damaged DNA (Hua, 1997). The length of the homology between the insert and the genome increases the specificity of targeting and cloning efficiency (Gray, 2001). Although there are several yeast integrating vectors that already exist (Sikorski, 1989), they have their own drawbacks. Namely, they do not comply by the BioBrick standards, they have short homologies with the yeast genome, they require special yeast deletion strains in order to function, and most do not provide colorimetric validation of successful integration. We therefore constructed a new plasmid (BBa_K319043) that is able to integrate into the ADE4 locus, knocking out the ADE4 ORF with high specificity. The ADE4 locus which is flanked by ATM1 and DYN3 was chosen as the site for integration because knocking out the ADE4 gene in an ADE2 mutant strain (such as YPH500) restores the white colony phenotype by undercutting the adenine biosynthesis pathway (Ugolini, 1996).
Figure 5: Adenine biosynthesis pathway retreived from http://www.phys.ksu.edu/gene/GENEFAQ.html. AIR gets converted into a red pigment. Mutations in ADE1 and ADE2 genes lead to the accumulation of the red colour in yeast. Mutations in upstream genes in the pathway do not allow for the generation of AIR, and therefore can restore the white phenotype in ADE1 and ADE2 mutants.
Therefore a white colony in this background indicates successful integration at the correct locus and eliminates the need for time consuming and costly PCR validation. BBa_K319043 is a BioBrick backbone that conforms to Assembly standard 23 (Phillips, 2006). The plasmid contains 200 bp homologous to the ATM1 terminator followed by the BioBrick cloning sites, followed by 200 bp homologous to the ADE4 terminator. Two version of the plasmid contain KanMX and NatMx expression cassettes (pADE4TAK and pADE4TAN respectively) between the ATM1 terminator and the BioBrick cut sites.
Results
Figure 1 shows a transformation into YPH500 of the ADE4 targeting vector with a natMX6 resistance cassette. 100% of the colonies are white, this means that in combination with the herterologous marker natMX6, the ADE4 targeting vector has a specificity of 100%. For comparison the following image shows a typical yeast transformation performed in our lab where a construct of interest in targeted to the ADE2 locus with Ura3 selection using 40 base pairs of homology added as an PCR primer overhang. Since the ADE2 locus is being targeted, red colonies indicate a successful transformation with targeting to the correct locus, and white colonies indicate targeting to the incorrect locus. Targeting to a specific locus in yeast is important because each locus has its own suceptipility to chomosomal positioning effects. If two constructs are to be compared, then this comparison must be performed in the same locus and the experimenter must ensure that the correct locus is targeted. 21 colonies are red and the other 64 colonies are white, this represents a 25% specificity. If loci that do not allow for colour selection are used, the experimenter must pick several colonies and screen them by PCR if only using 40 base pair homologies. This can be a tedious and costly process in high throughput applications such as library construction or screening and demonstrates why the ADE4 targeting vectors are useful.
Figure 6: A transformation of a construct of interest with URA3 selection into the ADE2 locus of BY4742 using 40 base pair overhangs. There are 21 red colonies and 64 white colonies.
Proximal – Distal promoter distinction
Ellis et al. developed a library of 40 variants of the inducible Gal1 promoter that contain operator sites for the binding of the TetR and the LacI repressors. These promoter libraries were generously donated to us. Eukaryotic promoters can be split into two halves, the proximal component and the distal component. The distal component is thought to be responsible for binding upstream activators. The proximal component is responsible for binding to the TATA binding protein and is directly involved in the recruitment of transcriptional machinery. We therefore hypothesized that it would be possible to shuffle around distal and proximal promoter components from various promoters. We proceeded to BioBrick the Act1 distal promoter and combine it with the proximal elements from the Ellis libraries. We were not able to finish this aspect of the project, however we have made all the BioBricks available to the registry. If this type of promoter shuffling in Eukaryotes is possible, then it can have a significant impact on eukaryotic synthetic biology. There are many interesting wild type promoters in nature that respond to a particular signal and these promoters are often used in biosensors. A problem with many of these interesting promoters is that although they respond correctly to a signal, they may not produced the desired output. By adding the correct proximal promoter, one should be able to tune the desired output rationally.
Characterization of the ADH1 promoter
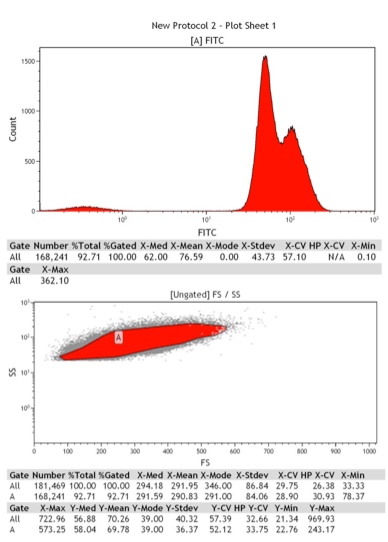
Although there are other versions of the ADH1 promoter already available in the registry, they are longer than needed for optimal expression from the promoter. Furthermore, since these promoters are longer, they are inhibited by ethanol which is used as a source of energy during the ethanol phase of the yeast life cycle. This information has been published by Ruohonen et al. We therefore decided to contribute an improved version of the ADH1 promoter in yeast. The length that was chosen for this ADH1 promoter is more manageable for the construction of larger networks, and the promoter is not inhibited by ethanol. We characterized the expression of the ADH1 promoter by cloning it in front of yEGFP with a ADH1 terminator. This construct was cloned into a pRS403 vector and integrated into the YPH500 genome. The resulting strain was innoculated in 3 mL of YPD + 2% Glucose + 2% Adenine overnight. The overnight culture was reinoculated into 2 mL of the same media at an OD of 0.02. After three hours the strain was analyzed on a Cyan ADP flow cytometer, 488 laser was used for exitation and emission was captured with a standard FITCH filterset. We observed roughly a 100 fold increase in GFP expression over wild type YPH500.
Results
Figure 7: Fluorescence of wild type YPH500
Figure 8: Fluorescence of ADH1 promoter driving yEGFP expression in a YPH500 background.
Power of yeast mating
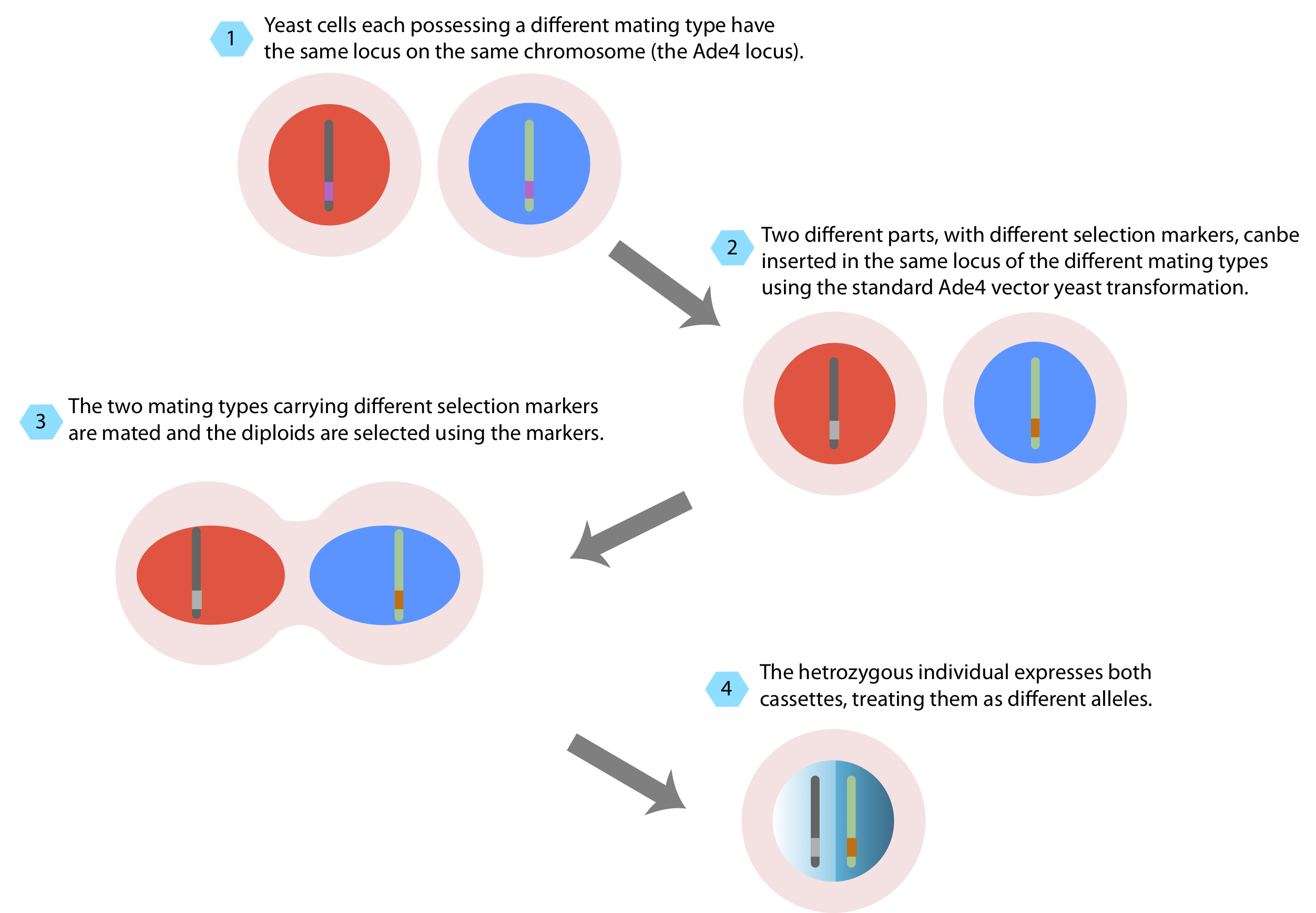
Yeast cells can live as hapoid organisms and as diploid organisms, the following schematic describes how two haploid yeast cells of different mating types that each contain part of a synthetic network can be combined to form a diploid yeast cell containing the full network.
Figure: 9 Schematic describing the yeast mating process
The two versions of the previously mentioned ADE4 targeting vector containing Nat and Kan selection are compatible with a yeast mating approach for combinatorial synthetic network construction. Half of a network can be cloned into the nat plasmid, and other half into the kan plasmid. The nat plasmid can be transformed into YPH499 and the kan plasmid can be transformed into YPH500, for example. If there are several different versions of the network in the nat plasmid (for instance each containing a certain promoter of different strength), it is much easier to mate them together than to clone them individually.
Possible network construction with our parts (Including toggle switches)
One of the goals of the 2010 iGEM team was to create a library of toggle switches. Although we did not manage to create this library we have contributed many of the BioBricks necessary for its construction. The rational behind choosing this project was not to generate the library, but to generate a toolkit of useful yeast components and use the library as a proof of principle. We have generated several useful BioBricks, and we hope they they will be used by the iGEM community!
References
- Guido, N. et al. A bottom-up approach to gene regulation. Nature 439, 856-860, doi:10.1038/nature04473 (2006).
- Hua, S.-B., Qiu, M., Chan, E., Zhu, L. & Luo, Y. Minimum length of sequence homology required for in vivo cloning by homologous recombination in yeast. Plasmid 38, 91-96 (1997).
- Gray, M. & Honigberg, S. M. Effect of chromosomal locus, GC content and length of homology on PCR-mediated targeted gene replacement in Saccharomyces. Nucleic Acids Research 29, 5156-5162 (2001).
- Sikorski, R. S. & Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27 (1989).
- Ugolini, S. & Bruschi, C. V. The red/white colony color assay in the yeast Saccharomyces cerevisiae: Epistatic growth advantage of white ade8-18, ade2 cells over red ade2 cells. Curr Genet 30, 485-492 (1996).
- Phillips, I. & Silver, P. A new biobrick assembly strategy designed for facile protein engineering. dspace.mit.edu (2006).
- Ellis et al. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol (2009) vol. 27 (5) pp. 465-71
- Ruohonen et al. Modifications to the ADH1 promoter of Saccharomyces cerevisiae for efficient production of heterologous proteins. Journal of Biotechnology (1995) vol. 39 (3) pp. 193-203
 "
"