Team:WashU/Notebook/MolecularBiology
From 2010.igem.org

2010/07/01
- Plated The Megax BH10B strain and Strain 8 from the cohen lab onto an amp plate to check that the Megax BH10B strain is killed
2010/07/13
- Minipreped all the cultures and nanodropped them
- Ran 3 PCR reactions (@ 58,60, and 62 C) with primers p1 and p2 in order to attach the kozak onto the YFP
- Enzyme digested Nat (bglII and EcoRI), Kan (BamHI and EcoRI), and Promoter (EcoRI). Did 2 reactions each with ~500 ng DNA
2010/07/14
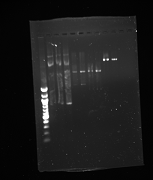
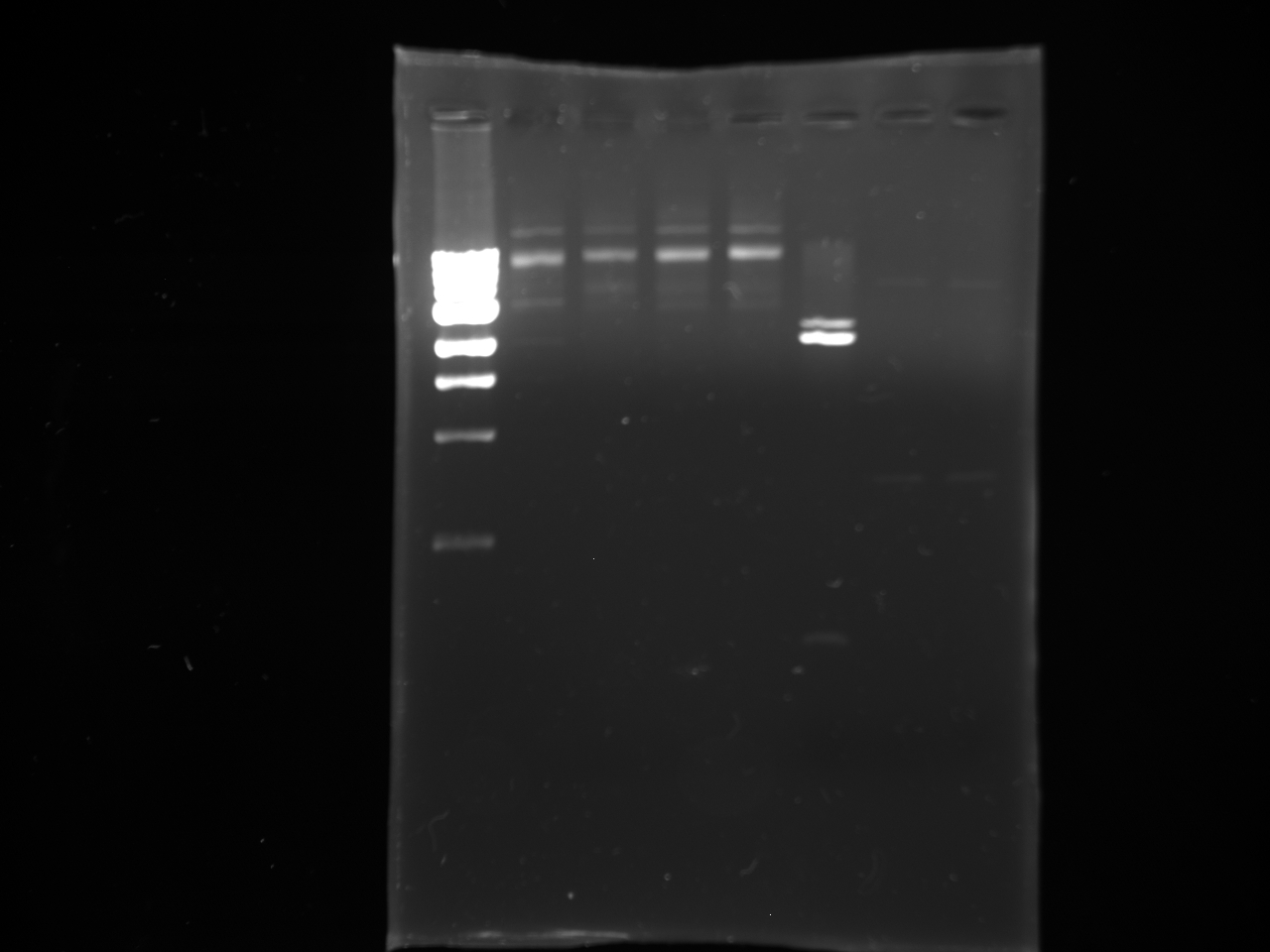
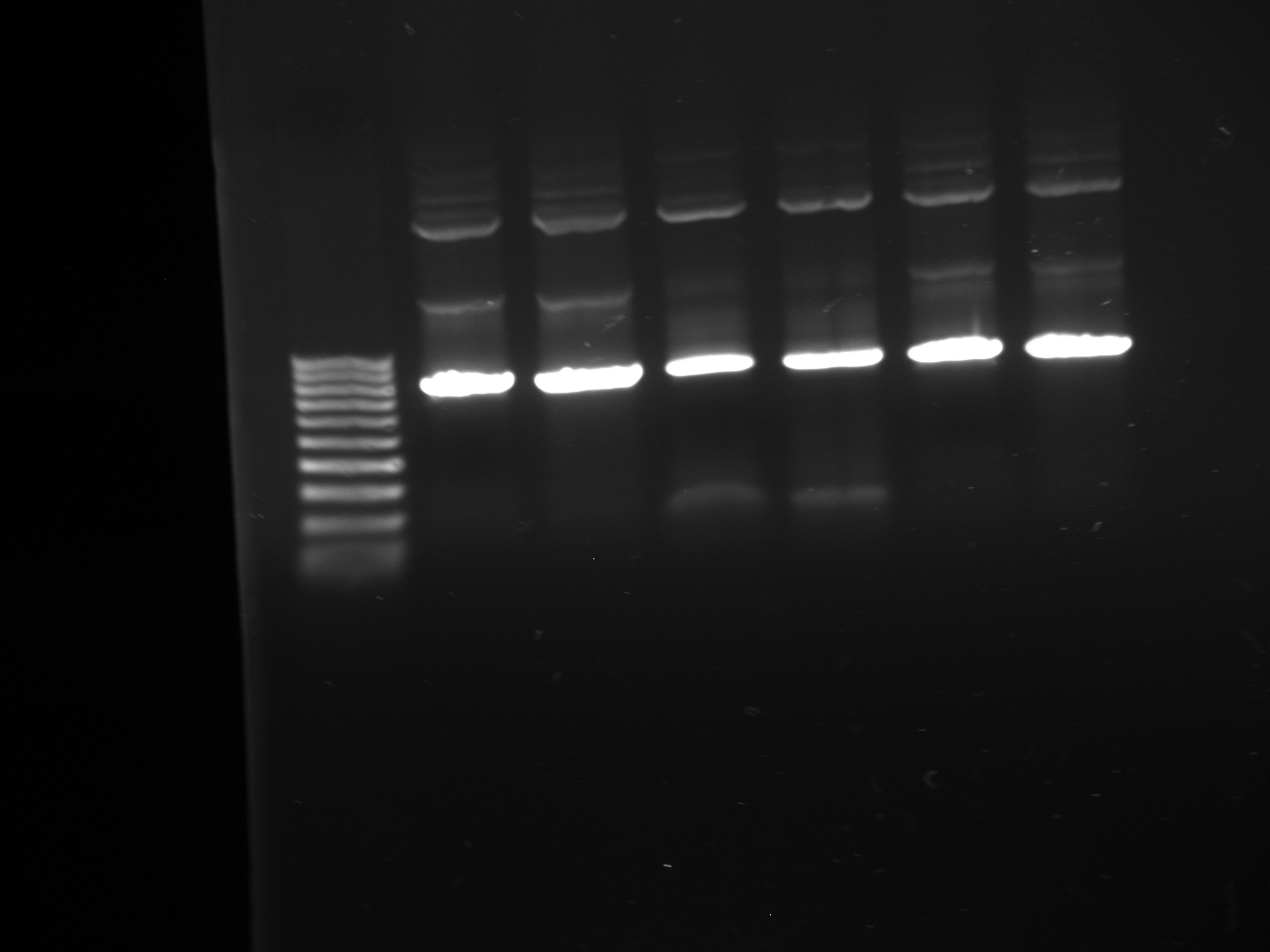
- Gel from July 13th
- Lanes from top to bottom:
1) Ladder 2) 5uL of 58 degree YFP + 1uL of loading dye 3) 5uL of 60 degree YFP + 1uL of loading dye 4) 5uL of 62 degree YFP + 1uL of loading dye 5) 5uL of KanMx4 1 + 1uL of loading dye 6) 5uL of KanMx4 2+ 1uL of loading dye 7) 5uL of NatMx4 1 + 1uL of loading dye 8) 5uL of NatMx4 2+ 1uL of loading dye 9) 5uL of Promoter 1 + 1uL of loading dye 10) 5uL of Promoter 2 + 1uL of loading dye
2010/07/15
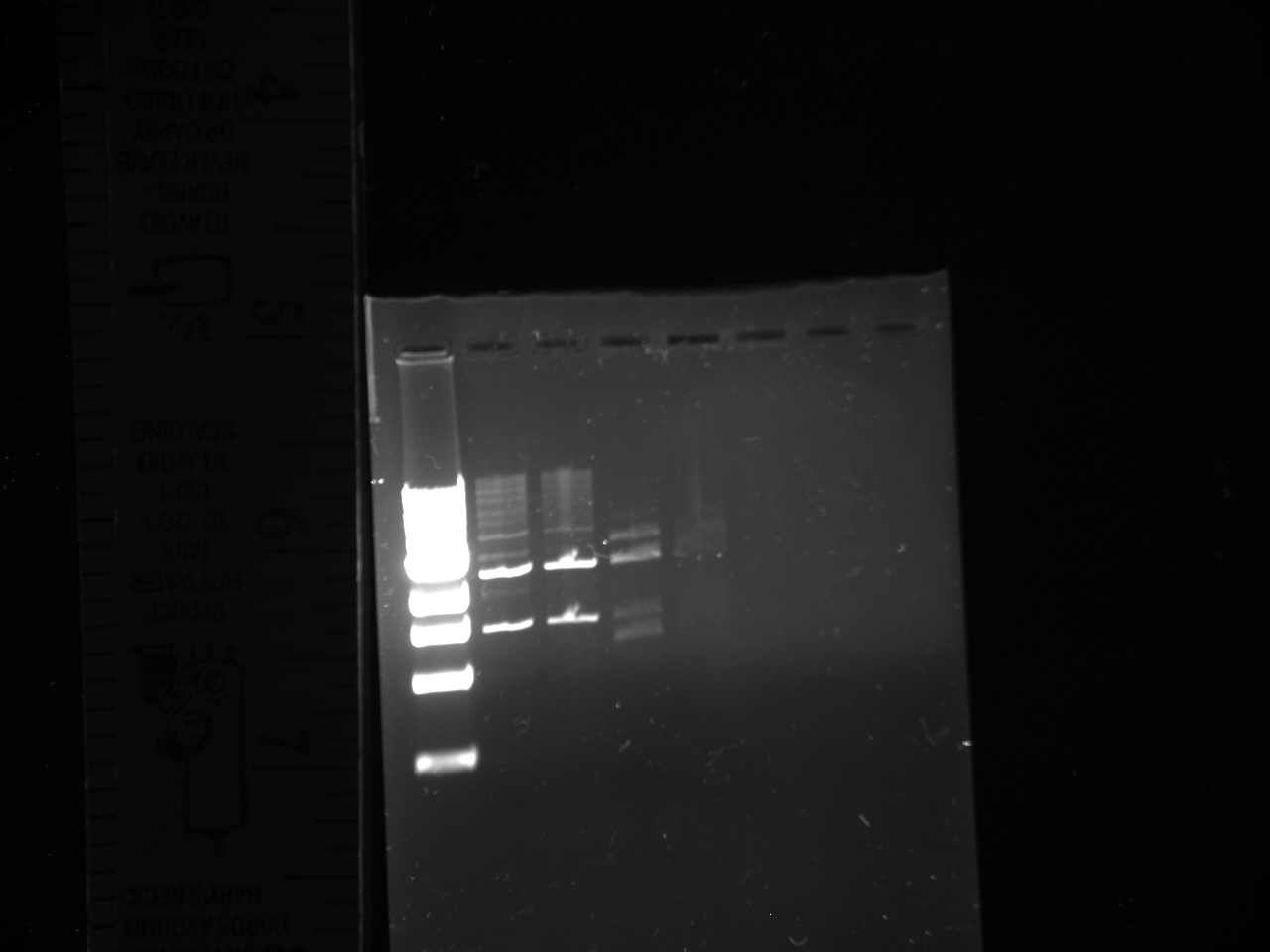
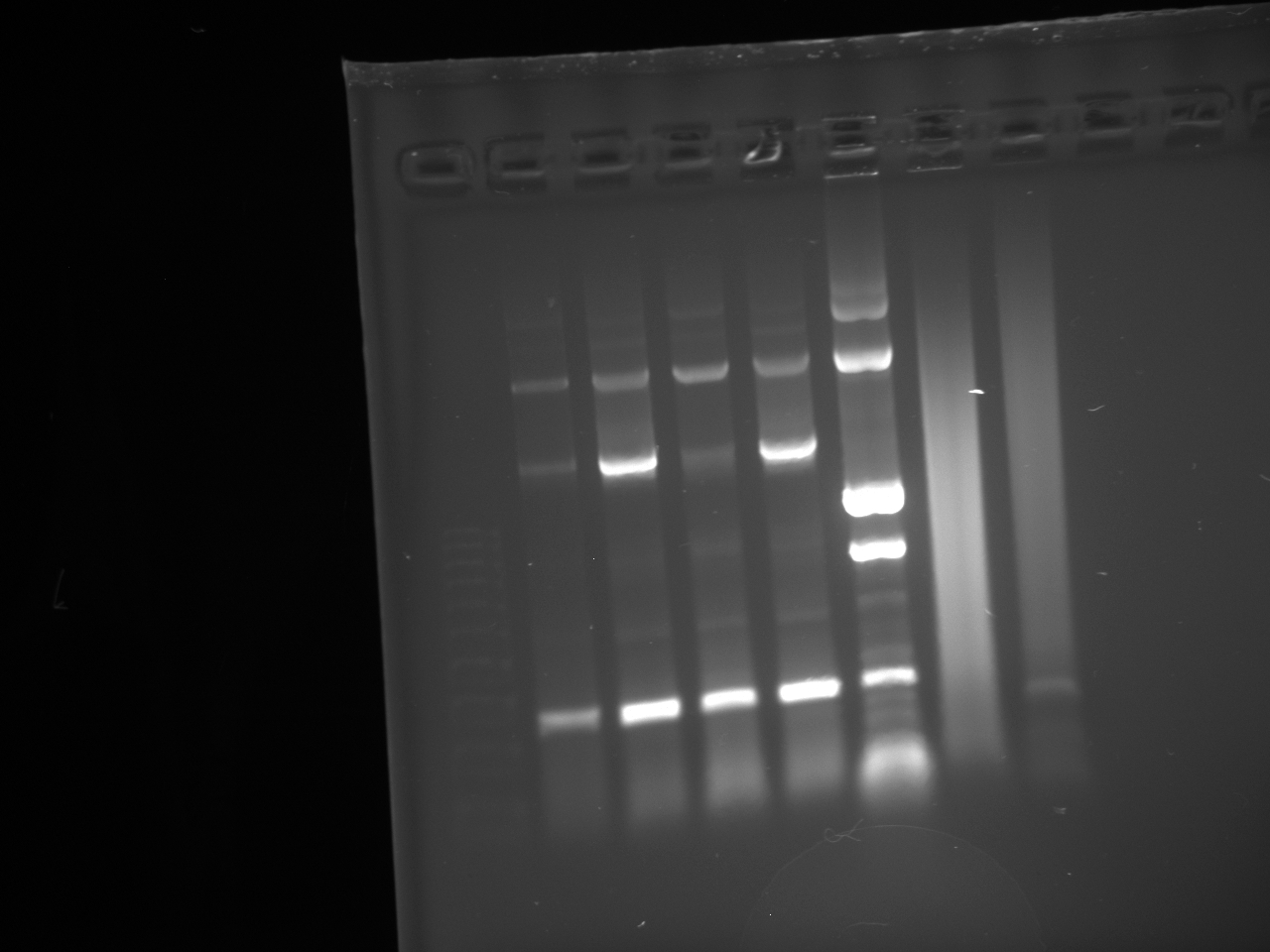
- Gel reads from left to right as follows:
Dna Ladder; NatMX4-Promoter construct 2x; KanMX4 plasmid backbone 2x.
- DNA ladder information is here at http://www.neb.com/nebecomm/products/productN3232.asp
2010/07/16
- Transformed EColi with the Kan plasmid containing the NatMX4 Promoter sequence.
- Bacteria Transformation July 16, 2010
1) Thaw competent cells on ice right before transformation. 2) Make 3 100uL tubes of competent cells 3) Add 4 ul of plasmid to cells, flick gently. 4) Incubate on ice 30 min. 5) Heat-shock cells for 30 seconds at 42oC w/o shaking 6) Transfer to ice, incubate 2 min 7) add 250 ul room temp SOC Media 8) Shake @ 37o for 1 hr 9) Spread 150 ul on a pre-warmed ampicilin selector plates, allow sample to dry on plate 10) Incubate o/n at 37oC upside down
2010/07/19
- Transformation of NatMX4/Promoter vector failed.
- Analysis of gel purification showed that the separate pieces which were ligated together on Thursday July 15th were of the correct length.
- Conclusion: the ligation failed; most likely due to denatured ligase. The ligase icebath had melted prematurely and heat inactivated the enzyme.
- Yeast group obtained several replacement ligases and necessary buffers from the Cohen Laboratory in the 4444 building of the WashU Med School.
- Transformation of C2 and SxL constructs into E. coli (AH)
- Reconstituted 5ug of each construct (in plasmid) in 10uL dH20. Final conc. of this stock solution is 500ng/uL. 10uL of working stock of each solution was made by diluting stock solution 1:10. Final conc. of working stock is 50ng/uL.
- Thaw 300uL tube of DH5alpha competent cells and divide among 5 tubes (60uL each). Amounts of 50ng/uL working stock were added to tubes labeled A-E in the following way:
A: 1uL C2 B: 5uL C2 C: 1uL SxL D: 5uL SxL E: Control=no DNA
- Incubate on ice 30 min, heat shock for 30s at 42C, rest on ice for 2 min.
- Add 250uL of LB to each tube, Shake at 37C for 1hr
- Plate 150uL on warm LB + Amp plates o/n at 37C
2010/07/20
- Digested 4 sequences:
1) NatMx4 with BglII and EcoRI: in microliters [5 buffer 3(NEB); 2.5 DNA at 200 ng/uL; 0.5 of BSA; 1 of each enzyme; 40 of H2O.] 2) Constitutive Promoter with EcoRI and SpeI: in uL [5 buffer 4(NEB); 2.5 DNA at 200 ng/uL; 0.5 of BSA; 1 of each enzyme; 40 of H2O.] 3) Construct 2 with BglII and XbaI: in uL [5 buffer 2(NEB); 1 DNA at 500 ng/uL; 0.5 of BSA; 1 of each enzyme; 41.5 of H2O.] 4) SxL with BamHI: in uL [5 buffer 3(NEB); 1 DNA at 500 ng/uL; 0.5 of BSA; 1 of BamHI; 41.5 of H2O.]
- all were incubated for 4 hours followed 20 minutes of heat inactivation.
2010/07/21
- Digest SxL construct with EcoRI: added 1uL EcoRI to already done mix from 20 July digestion.
- Gel purify NatMx4, Promoter, Construct 2, and KanMx4 digest results from 20 July 2010
Note: KanMx4 bands were not observed but approximate region of gel was removed for ligation KSB.
- Ligate all three pieces together: NatMx4, Promoter, and Construct 2
- 2 reactions:
1) NCPA = NatMx4+Promoter+Construct 2
3.5uL Nat + 3.5uL Pro + 1.5uL C2
2) NCPB = NatMx4&Promoter+Construct 2
[NatMx4&Promoter was pre-existing ligation product from 15/16 Jul]
7uL NP + 1.5uL C2
- Ligate KanMx4 with Sxl
- 2 reactions:
1) KSA = KanMx4 Digest 2 + SxL
7uL KanDigest2 + 1.5uL SxL
2) KSB = KanMx4 (purified) + SxL
7uL GelPureKanMx4 + 1.5uL SxL
- Freezer stocks of C2 and SxL
- 1:1 ratio of LB was mixed with 100% glycerol to make a 50% glycerol/LB solution.
- 750uL of glycerol/LB solution was added to 750uL of each C2 and SxL culture from transformation on 2010/07/19.
- Two tubes of each were placed at -80C.
2010/07/22
- Transformed the ligation DNA from reactions NCPA, NCPB, KSA, and KSB into E. Coli using the following protocol.
- Bacteria Transformation
1. Thaw competent cells on ice right before transformation. 2. Add 4ul of plasmid to cells, flick gently. 3. Incubate on ice 30 min. 4. Heat-shock cells for 30 seconds at 42oC w/o shaking 5. Transfer to ice, incubate 2 min 6. add 125 ul room temp LB Media 7. Shake @ 37o for 1 hr + 8. Spread 100 ul on a pre-warmed plate w/ correct antibiotic selector, allow sample to dry on plate 9. Incubate o/n at 37oC upside down
Week of 7/26
7/26
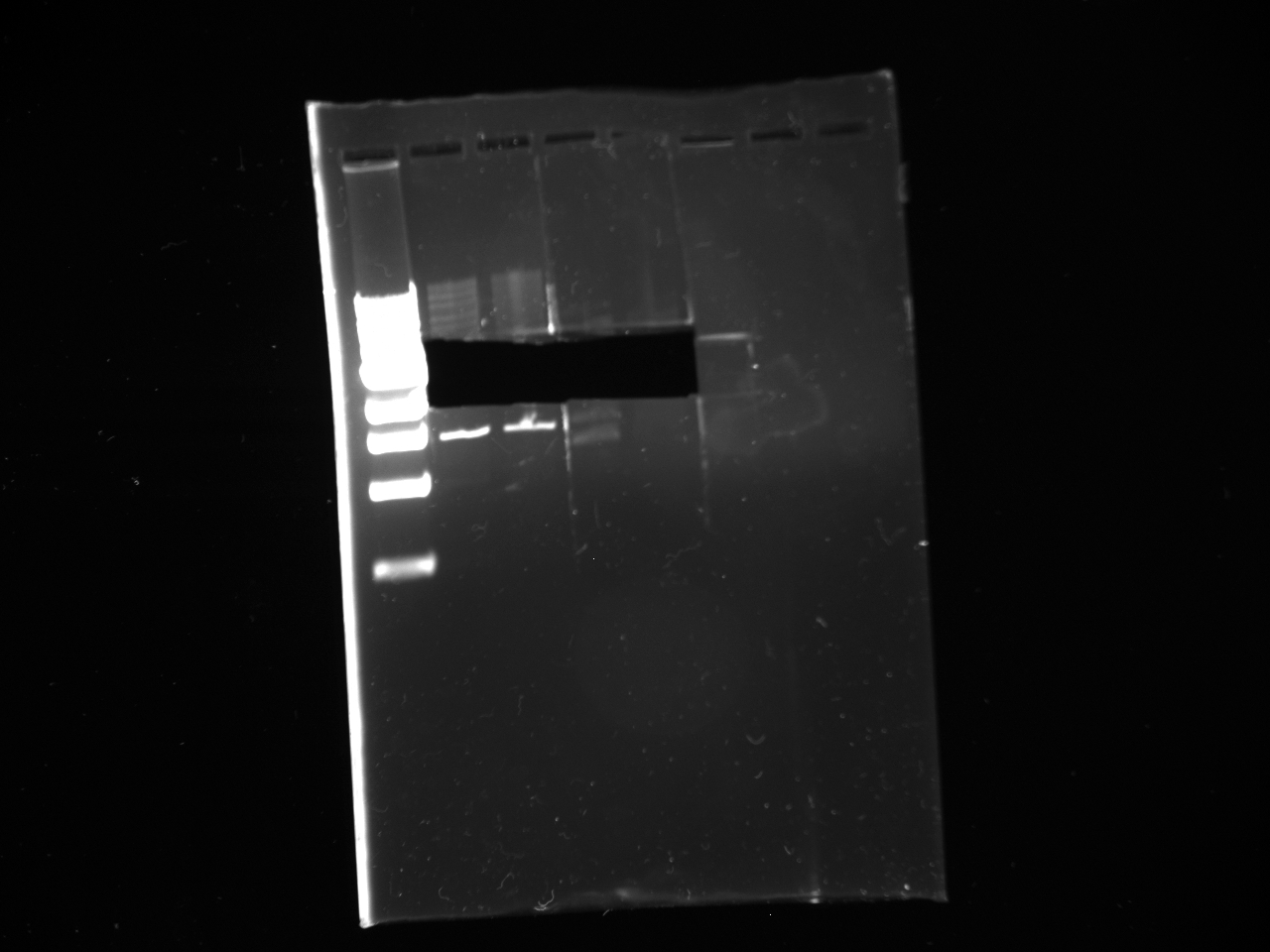
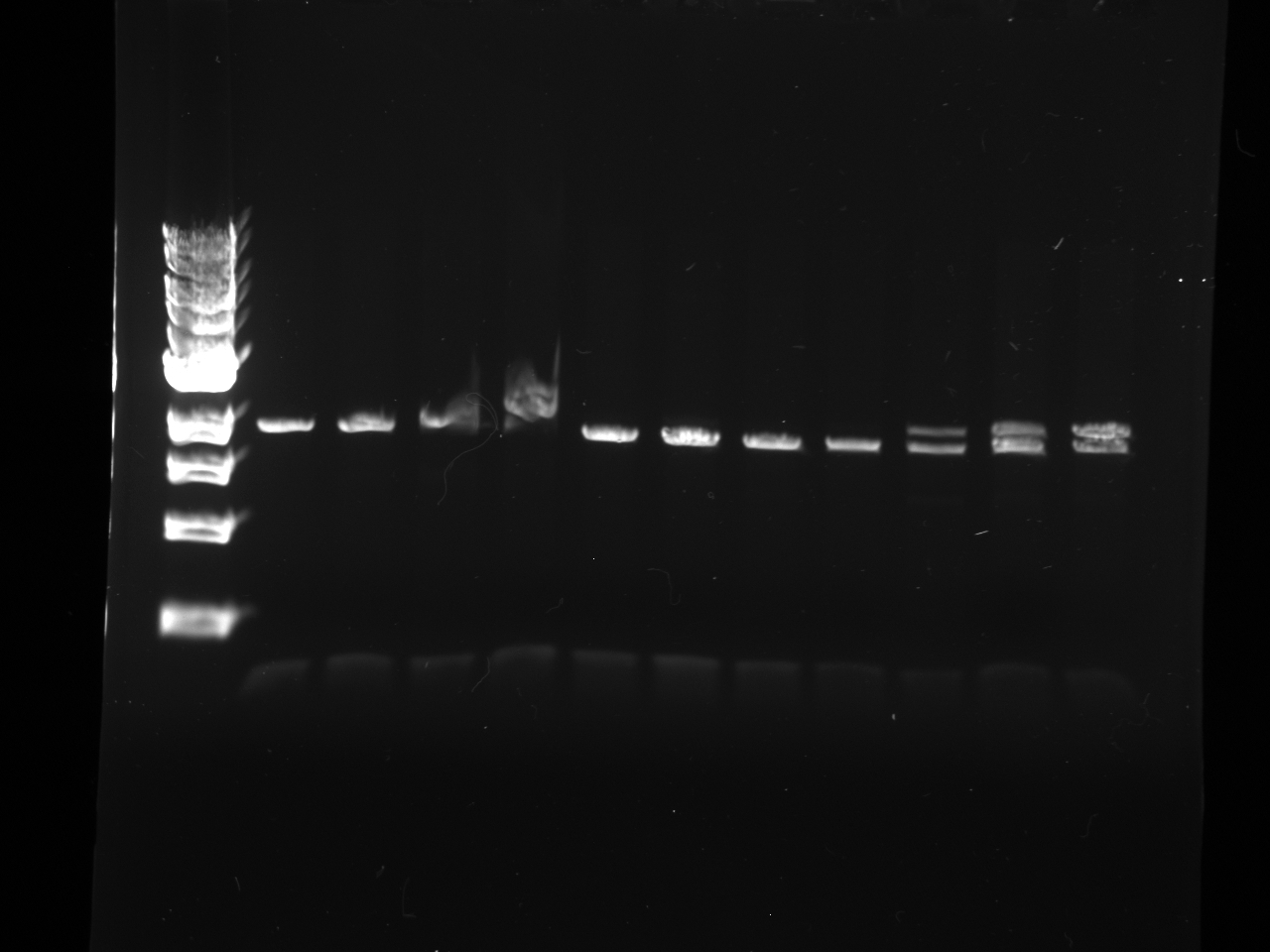
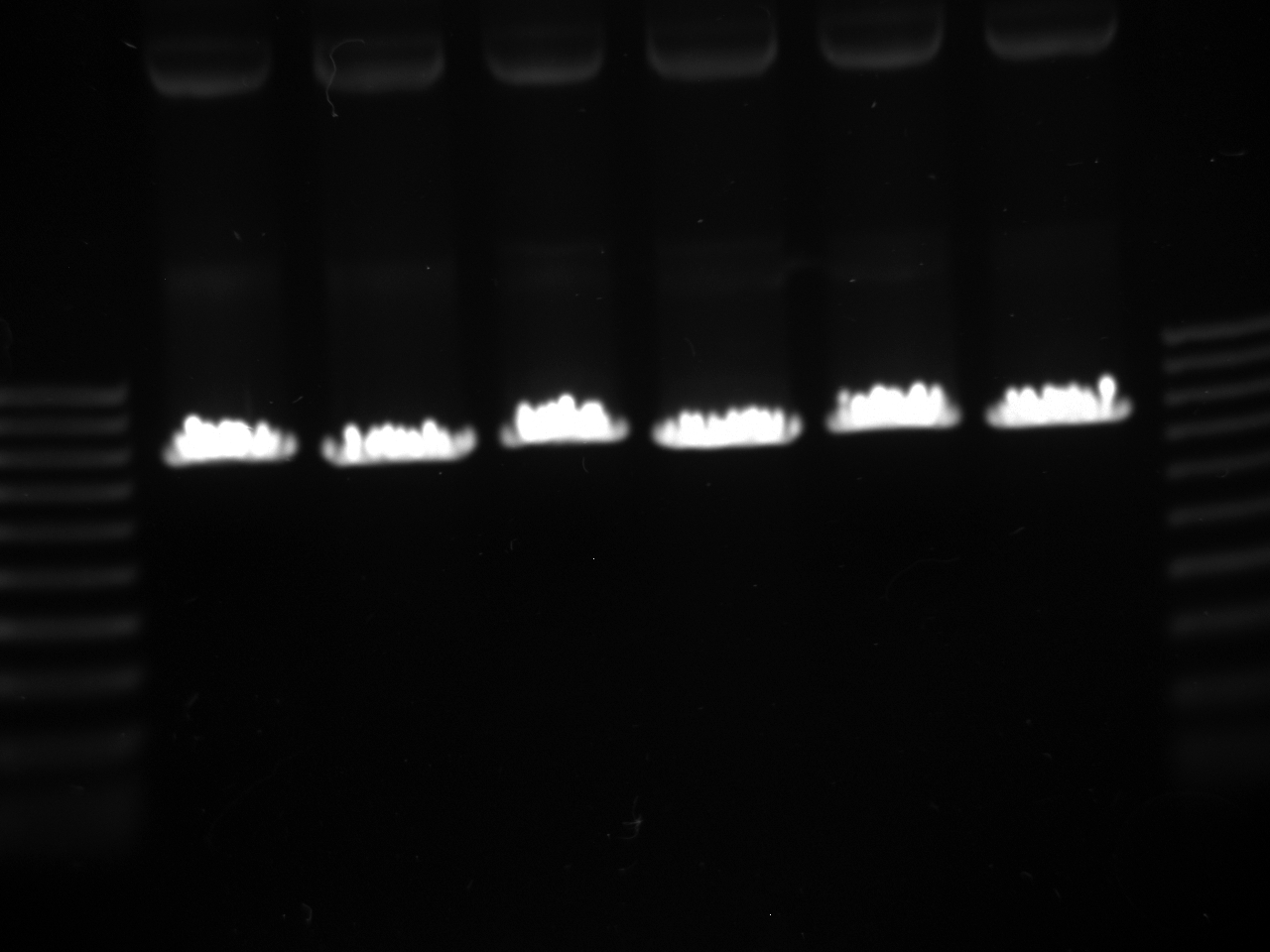
Mol. Bio. Group Digest and gel purify to confirm the results of the SxL construct transformants created on July 22. Gel image is attached.
Reactions 4, 5, 6, and 7 of the Yeast Group's gel are from July 21 ligation, July 22 transformation, and the July 24 miniprep of the July 23 cultures. [The leftmost lane is a ladder; numbering begins at the second lane with Number 1.]
Sequencing the YFP to confirm the Kozak Sequence:
iGEM1 YFP 62 degrees C primer P9
iGEM2 YFP 62 degrees C primer P2
iGEM3 YFP 62 degrees C primer P9
iGEM4 YFP 62 degrees C primer P2
7/27
Digestion of Terminator and YFP pieces (YFP is currently being sequenced to confirm Kozak Sequence)
Terminator
• 37.5 uL of H20 • 5 uL of NEB buffer 3 • 5 uL of Terminator DNA • 1 uL of Pst1 enzyme • 1 uL of Xba1 enzyme • 0.5 uL of BSA
YFP
• 37.5 uL of H20 • 5 uL of NEB buffer 2 • 5 uL of YFP DNA • 1 uL of Pst1 enzyme • 1 uL of Spe1 enzyme • 0.5 uL of BSA
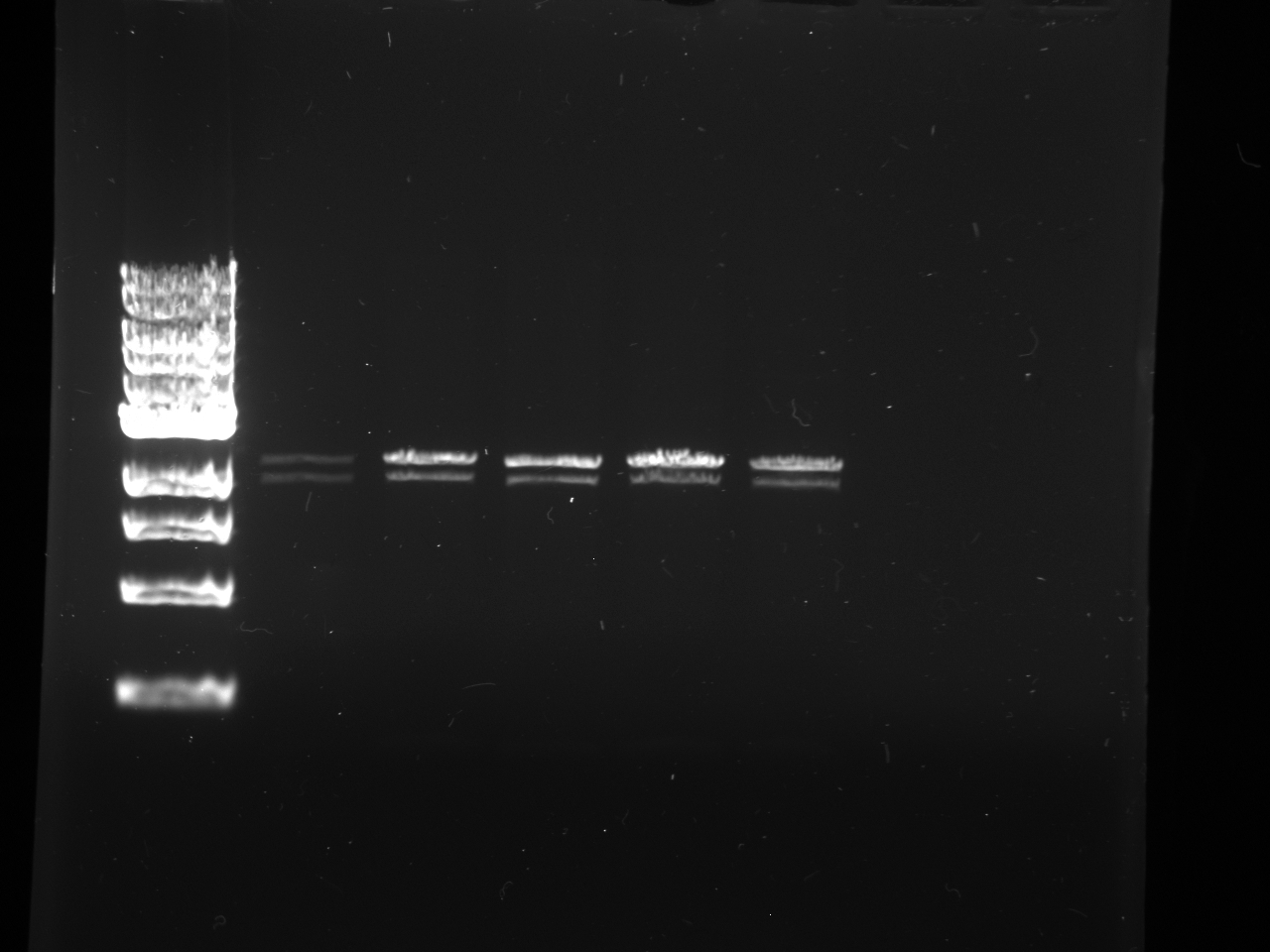
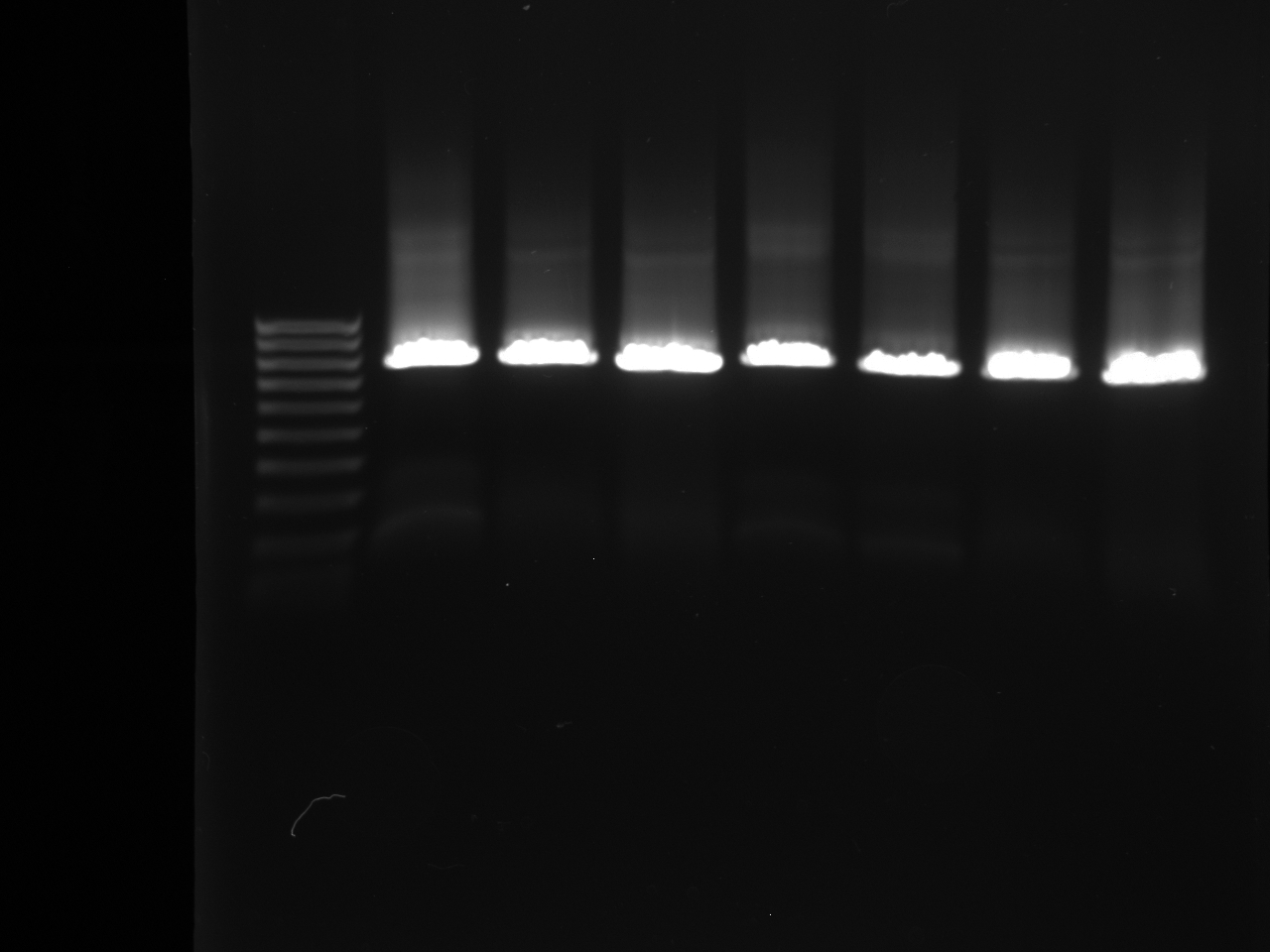
Gel Purification and Confirmation of NPCA, NPCB, Terminator, and YFP
Gel Lanes: 1) Ladder. 2) NPCA 1. 3) NPCA 2. 4) NPCB 1. 5) NPCB 2. 6) Terminator. 7) YFP. 8) YFP.
7/28
Double Digests of KSA2 (confirmed); NPCA 1 & 2; and NPCB 1 & 2.
• 32.5 uL of H2O • 10 uL of DNA • 5 uL of NEB buffer 4 • 1 uL of XhoI • 1 uL of AvrII • 0.5 uL of BSA
Began Colony PCR of NPCA and NPCB plates: 14 colonies taken from each.
7/29
Colony PCR Primers p1 and p5 are defective Colony PCRs will not yield proper results
The back up cultures still are intact and can be reused when the proper primers arrive
Digestions:
YFP Biobrick:
• 32.5 uL of H2O • 5 uL of DNA • 5 uL NEB buffer 3 • 5 ul CIP • 1 uL PstI • 1 uL XbaI • 0.5 uL BSA
YFP kozak [Mix is defective due to Primer 1 error; DNA does not contain XbaI site]
• 30 uL kYFP DNA • 12.5 uL of H2O • 5 uL NEB buffer 4 • 1 uL XbaI • 1 uL SpeI • 0.5 uL BSA
C2 control
• 37.5 uL H2O • 5 uL DNA • 5 uL NEB buffer 4 • 1 uL XhoI • 1 uL AvrII • 0.5 uL BSA
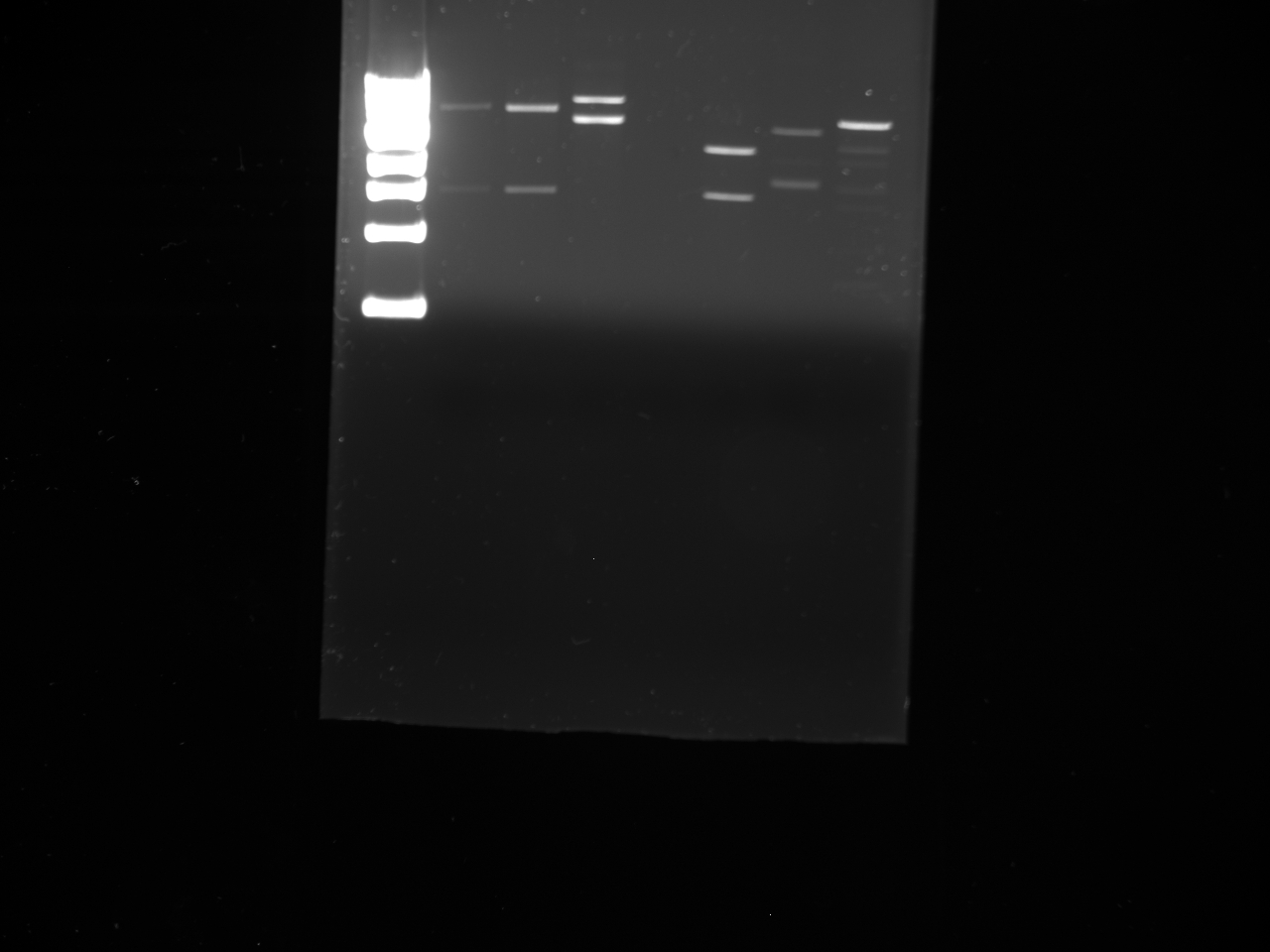
Gel:
• Lane 1: Ladder 1kb • Lane 2: C2 control • Lane 3: NPCA1 3.7 and 2.3kb • Lane 4: NPCA2 3.7 and 2.3kb • Lane 5: NPCb1 3.7 and 2.3kb • Lane 6: NPCb2 3.7 and 2.3k
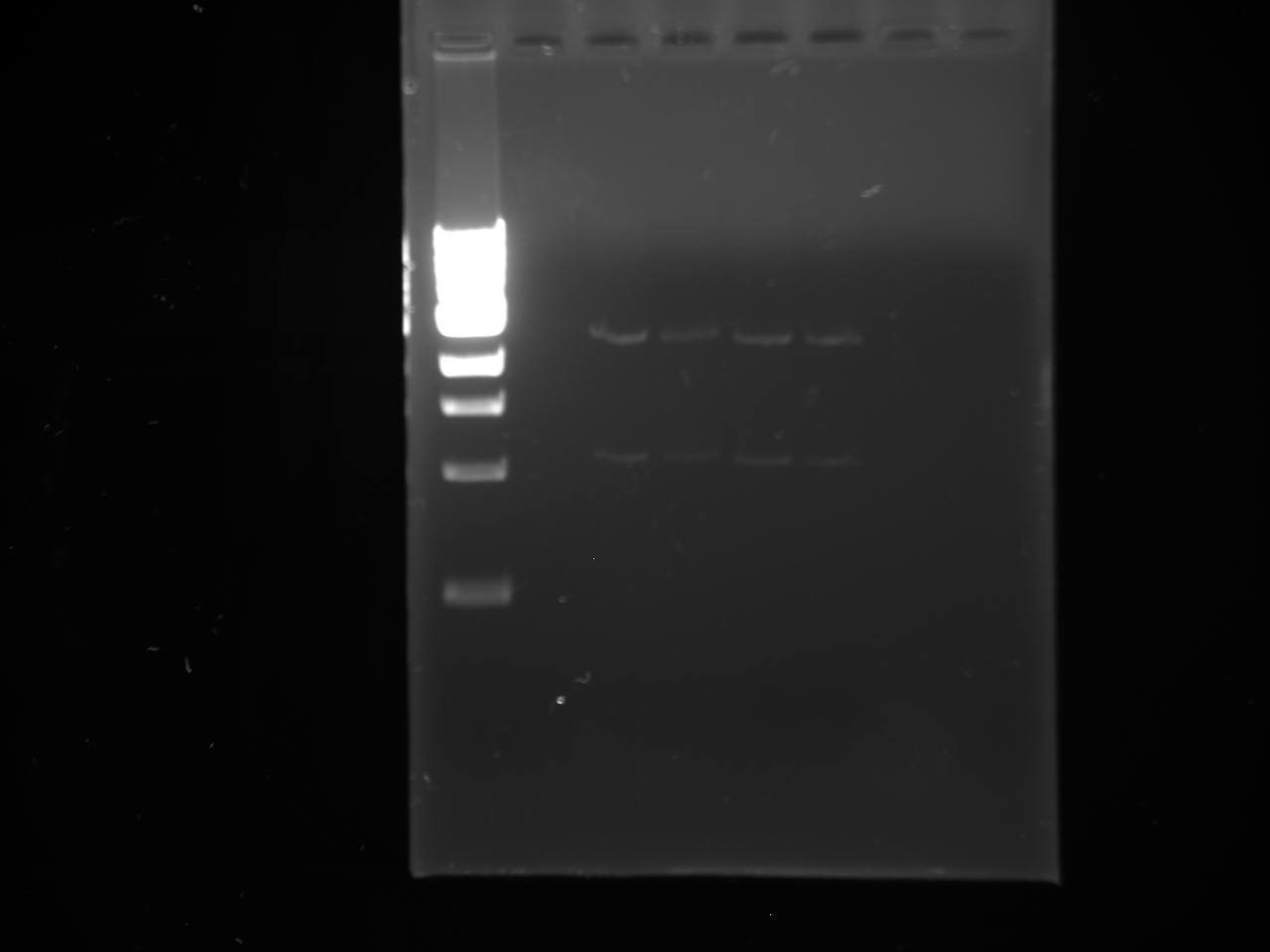 Conclusion NP was not present within the construct 2 plasmid
Conclusion NP was not present within the construct 2 plasmid
Gel 2:
YFP backbone--- not observed Gel tossed out
Ligations:
NPCc: ligate C2// BglII & XbaI with CIP A with NatMx4 and Promoter
• 1uL Y4 ligase buffer • 0.5 uL ligase • 3 uL NatMx4 • 3 uL Promoter • 2.5 uL C2
NPb: NatMX4 ligates to Promoter
• 17 uL NatMx4 • 17 uL Promoter • 4 uL T4 ligase buffer • 2 uL ligase
Extra Digestion:
C2 // bglII & XbaI CIP B
• 27.5 uL H2O • 10 uL DNA • 5 uL CIP • 5 uL NEB buffer 3 • 1 uL BglII • 1 uL XbaI • 0.5 uL BSA
7/30
Gel purify 30 uL NPb and 30 uL C2b
Make NPCd: Ligate NPb & C2b (non purified) ~ 10 uL of each
• 5.5 uL NPb • 3 uL C2b • 0.5 ligase • 1 uL buffer
Make NPCe: Ligate NPb & C2b (purified)
• 24 uL of NPb DNA • 1.5 uL of C2b DNA • 3 uL buffer • 1.5 uL ligase
Re-attempt ligation of NatMx4 and Promoter
• 25.5 uL mix of NatMx4 and Promoter • 3 uL buffer • 1.5 uL ligase
Week of 8/2
8/2
Transformations:
NPCc NPCd NPCe and KSA2
1. Thaw competent E.Coli cells on ice right before transformation. 2. Make 4 tubes with 70uL of competent cells each 3. Add 3 ul of NPCc, d and e plasmid to 3 tubes and 1 uL of KSA2 to the last tube, flick gently. 4. Incubate on ice 30 min. 5. Heat-shock cells for 30 seconds at 42oC w/o shaking 6. Transfer to ice, incubate 2 min 7. add 140 ul room temp LB Media 8. Shake @ 37o for 2 hr [to ensure recovery from shock] 9. Spread 100 ul on a pre-warmed ampicilin selector plates, allow sample to dry on plate 10. Incubate o/n at 37oC upside down
PCR reactions:
Reaction 1st round
kYFPa: 94ng of YFP at 59 degrees C using p10 & p2
kYFPb: 94ng of YFP at 61 degrees C using p10 & p2 ---------------------------------------------------
Reaction 2nd Round
kYFPa1: 1 uL of kYFPa at 59 degrees C using p11 & p2
kYFPb1: 1 uL of kYFPb at 59 degrees C using p11 & p2
kYFPa2: 1 uL of kYFPa at 61 degrees C using p11 & p2
kYFPb2: 1 uL of kYFPb at 61 degrees C using p11 & p2
8/3
Cultures:
Created liquid cultures from the transformation results of Ksa2, NPCc, NPCd, NPCe, and C2.
[C2 transformation was performed by Amanda when the C2 construct arrived]
Digests:
kYFPa1, a2, b1, b2 and YFPBiobrick on backbone pSB1C3
For ALL:
• 27.5 uL of H2O • 15 uL of DNA of the respective construct • 5 uL of NEB buffer 3 • 1 uL of PstI enzyme • 1 uL of XbaI enzyme • 0.5 uL of BSA
[resulting products were column purified and eluted to 50 uL]
Ligation of kYFP's and the backbone
• 21 uL of kYFPa1, a2, b1, b2 in 4 respective tubes • 4.5 uL of BB into all four tubes • 3 uL buffer • 1.5 uL T4 ligase
Of NatMx4 and Promoter
NatMx4
• 35 uL of H2O • 7.5 uL of DNA • 5 uL of NEB buffer 3 • 1 uL of bglII • 1 uL of EcoRI • 0.5 uL BSA
Promoter
• 35 uL of H2O • 7.5 uL of DNA • 5 uL of NEB buffer 4 • 1 uL of EcoRI • 1 ul of SpeI • 0.5 uL BSA
8/4
Gel purification of NatMX4 and Promoter
4 lanes Nat and 4 lanes Pro
Successfully extracted the lower of 2 visible lines from all lanes [top line was vector; bottom was Nat and Pro inserts]
Miniprep
Kan-SxL construct and Construct 2
Transformation
kYFPa1, a2, b1, b2 on the Biobrick Backbone.
1. Thaw competent cells on ice right before transformation. 2. Make 4 75uL tubes of competent cells 3. Add 4 ul of plasmid to cells, flick gently. 4. Incubate on ice 30 min. 5. Heat-shock cells for 30 seconds at 42oC w/o shaking 6. Transfer to ice, incubate 2 min 7. add 150 ul room temp LB Media 8. Shake @ 37o for 1 hr 9. Spread 100 ul on a pre-warmed ampicilin selector plates, allow sample to dry on plate 10. Incubate o/n at 37oC upside down
Ligation
NatMx4 + Promoter
• 30 uL of DNA (mix of both parts; they were extracted together) • 4 uL of Buffer • 4 uL of H2O • 2 uL of T4 ligase
8/6
Transformation
kYFP on the Biobrick Backbone.
1. Thaw competent cells on ice right before transformation. 2. Make 1 100 uL tubes of competent cells 3. Add 4 ul of plasmid to cells, flick gently. 4. Incubate on ice 30 min. (10:45 to 11:15) 5. Heat-shock cells for 30 seconds at 42oC w/o shaking 6. Transfer to ice, incubate 2 min 7. add ul room temp SOC Media 8. Shake @ 37o for 1 hr 9. Spread 100 ul on a pre-warmed ampicilin selector plates, allow sample to dry on plate 10. Incubate o/n at 37oC upside down
Week of 8/9
8/9
Make Tetracycline plates to select for kYFP transformants
Transform kYFP into E.Coli
1. Thaw competent cells on ice right before transformation. 2. Make a 75 uL tube of competent cells 3. Add 2 ul of plasmid to cells, flick gently. 4. Incubate on ice 30 min. (12:10 to 12:40) 5. Heat-shock cells for 30 seconds at 42oC w/o shaking 6. Transfer to ice, incubate 2 min 7. add 125 ul room temp SOC Media 8. Shake @ 37o for 1 hr 9. Spread 100 ul on a pre-warmed tetracycline selector plates, allow sample to dry on plate 10. Incubate o/n at 37oC upside down
8/10
Made a culture from possible colonies on tetracycline plate
Double Digest of pSB1A3
• H2O • DNA • NEB buffer 3 • XbaI • PstI • BSA
Gel purification of digest products
1kb ladder, 3 lanes of pSB1A3, and 3 lanes of YFP-BB
results: all 3 bands observed (one for the pSB1A3 digest and 2 for the YFPbb) [an extra band from the biobrick was seen above the 2 desired YFP bands possibly single or uncut plasmid]
Gel extraction of digest products
1 extraction for each of the 3 desired bands
Ligations
• kYFP to pSB1A3 • kYFP to Biobrick backbone • YFP to pSB1A3 • YFP to Biobrick backbone
Transformation of the ligation products into E.Coli
1. Thaw competent cells on ice right before transformation. 2. Make 4, 70 uL tube of competent cells 3. Add 2 l of plasmid to cells, flick gently. 4. Incubate on ice 30 min. (3:45 to 4:15) 5. Heat-shock cells for 30 seconds at 42oC w/o shaking 6. Transfer to ice, incubate 2 min 7. add 125 l room temp SOC Media 8. Shake @ 37o for 1 hr 9. Spread 100 l on a pre-warmed ampicilin selector plates, allow sample to dry on plate 10. Incubate o/n at 37oC upside down
8/11
Colony PCR
• 4 possible transformant colonies PCR'd and back-up culture made
PCR
• Re-PCRing the kYFP at 59, 60, 62 degrees C [product was named kYFP2]
Transformation of kYFP with o/n ligation results
• 50uL of cells • 2 uL of kYFP on Biobrick backbone • 2 uL of kYFP on pSB1A3 • 150uL of SOC media • plated all of the incubated/shaken culture
Gel purifications
100b ladder; 4 lanes of colony PCR; 59*C PCR of new kYFP; 60*C PCR of new kYFP; 62*C PCR of new kYFP.
results
 Digests of PCR Products and the Terminator
Digests of PCR Products and the Terminator
Digesting PCR kYFP with XbaI and SpeI
Digesting terminator [in vector] with XbaI and CIP
Ligation
8/12
Miniprepped the cultures made on the 11th [#4 had no growth or pellet]
Created cultures of the 4 transformant colonies from the 11th's transformation
redesigned the kYFP reverse primer
Ligate Terminator and kYFP2 (from the 11th's digestion)
[30min room temp ligation]
PCR at 60*C the Miniprep results, a YFP control and the Ligation product kYFP2term with primers p2 and p12& p9 and p12 reactions with the YFP control and the kYPF2term. [7 rxns total as #4 yielded no pellet for miniprepping]
[p2 was the questionable reverse kYFP primer]
Gel run: 100bp ladder, culture 1 DNA; cult. 2 DNA; cult. 3 DNA; YFP 2-12 PCR; YFP 9-12 PCR; kYFP2 2-12 PCR; kYFP2 9-12 PCR
results
Conclusion: Colonies were not desired kYFP transformants [match the pattern of the YFP (2-12) control]
Transformation of E.Coli with the kYFP2term plasmid (ligation product)
3uL in 50uL of cells 150uL SOC media 75 ul plated onto amp selector plates
8/13
Transformation yielded a large number of colonies which may contain the kYFP2term construct
pour new ampicilin plates
Beginning screening processes today:
Grow 16 cultures of several colonies from transformants
Future screening steps:
Miniprep the cultures
digest DNA with XbaI & PstI
digest C2 with XbaI & PstI
Gel12 purify C2 and the correct kYFP2term from the improper kYFP2term [X will cut improper into 3 pieces, correct into 2]
30min room temp ligate correct kYFP2term with C2
Transform the ligated construct, C2kYFP2term, into E.Coli
Grow cultures from transformation
Miniprep the cultures
submit DNA for sequencing
digest with bglII & XbaI & CIP
Gel purify
30 min room temp ligation of C2kYFP2term with the NatMX4-Promoter construct
Transform ligation product into E.Coli
8/14
Miniprepped the cultures (16) made from kYFP2term transformants
Digests
Digested C2 with SpeI and PstI and CIP in NEBuffer 2. [27.5 H2O; 14 DNA 1 CIP; 1 SpeI; 1 PstI; 5 buffer; 0.5 BSA]
6 Digests of Miniprepped DNA with XbaI and PstI in NEBuffer 2 [35.5 H2O; 7 DNA; 5 buffer; 1 XbaI; 1 PstI, 0.5 BSA]
Note: rxns 12,13,14,15, and 16 used low concentration XbaI
Gels:
12 lane gel contained: 1kb ladder; culture samples 1-11
8 lane gel contained: 1 kb ladder; culture samples 12-16
Expected results
1. Successful construction: 2 kb band and .993kb band 2. Backwards kYFP2: 3 kb band 3. No kYFP2: 2kb band and .25 kb band
results
Conlcusion: #3 results for all samples; no kYFP2 ligated into the terminator
Week of 8/16
8/19
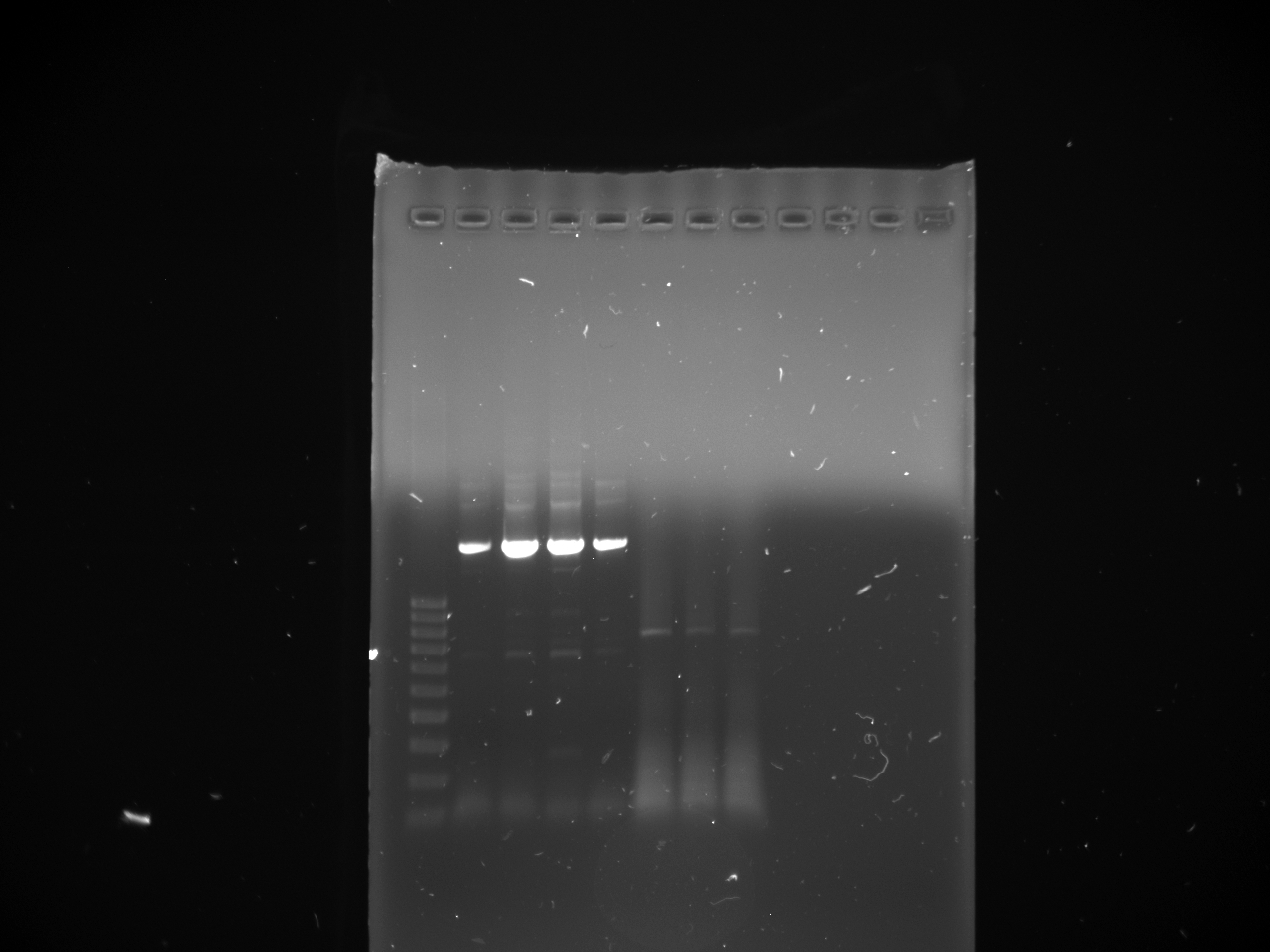
Got the new primers in today. Ran a PCR to make the new KYFP inserts. Used 1 ul of YFP biobrick plasmid as the template. p15 and p 16 were used to make the KYFP3. p10 and p17 were used to make the preKYFP4. Three PCR reactions were set up for each and were then run at 58, 60, and 64 C with 1 min extension time. The PCR products were then run out on a gel. The bands coresponding to ~ 800 bp were cut out and gel purified.
lane
1 100 bp ladder 2,3 KYFP3 @58 4,5 KYFP3 @60 6,7 KYFP3 @64
lane 1 100 bp ladder 2,3 preKYFP4 @58 4,5 preKYFP4 @60 6,7 preKYFP4 @64 8 100 bp ladder
The preKYFP4 products were cleaner as was to be expected. The KYFP3 @60 has anomalous lower bands ~200 bp and the KYFP3's in general have more minor banding patterns
Running a PCR over night.
• 1 ul of KFYP3 64+p13+p16=KYFP5 o running 2 reactions: one at 60 and the other at 62 C • 1 ul of preKYFP4 64+p11+p16=KYFP4 o running 2 reactions: one at 60 and the other at 62 C
8/20
Molecular Biology Group
Ran the PCR products out on a gel.
Lane
1 100 bp ladder 2 KYFP5 @60 3,4 KYFP4 @60 5,6 KYFP5 @62 7,8 KYFP4 @62
The bands that are ~800 bp were cut out and purified. Then I set up a five hour enzyme digest @ 37 C. I cut KYFP3 @64 w/ XbaI and PstI, KYFP4 @62 w/ XbaI and PstI, KYFP5 @62 w/ XbaI and PstI, YFP backbone w/ XbaI, PstI, and CIP, and Terminator with XbaI and CIP. The digested results were run out on a gel and then the appropriate bands were excised and the DNA was purified out. An over night RT ligation was set up with 1 ul of backbone: 7.5 ul insert. Cut KYFP3, KYFP4, and KYFP5 were ligated into the cut YFP back bone. A negative control (only cut YFP back) and a positive control (YFP insert back into YFP back bone) were also set up.
8/21
The ligation reactions (KFYP3, KYFP4,KYFP5, - control, + control) were transformed and plated onto amp plates.
Biobrick Group Transformation Results
Plate Result 1. K58&p1 2 colonies formed, one on the edge of the plate pointing towards a possible satellite colony 2. N58&p1 No colonies appeared to have formed 3. K60&p1 No colonies appeared to have formed 4. N60&p1 No colonies appeared to have formed 5. K64 &p1 No colonies appeared to have formed 6. N64 & p1 No colonies appeared to have formed pSB1AT3 Many ~100 colonies formed with over 50% expressing red color under room light, predominantly large colonies are red pSB1C3 ~30 colonies formed with slght red coloring at the center
Over Night Colonies prepared from
K58&p1 colony from middle of plate 34μg/ml chloramphenicol liquid media pSB1AT3 50μg/ml Amp liquid media pSB1C3 34μg/ml chloramphenicol liquid media
Week of 8/23
8/26
Results from transformation on 8/25
Number Plasmid Plate Observations 1 pSB1AT3 Tet Lawn of colonies with sprinkled red transformants 2 None Tet Lawn of colonies 3 pSB1AT3 Amp ~500 colonies, all small, mostly red transformations but some 4 None Amp ~100 colonies, some very large in size 5 pSB1K3 Kan ~80 colonies observed all red transformants 6 None Kan No colonies observed
 "
"