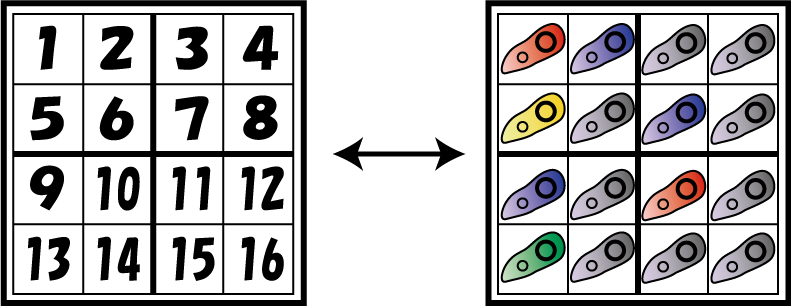Team:UT-Tokyo/Sudoku abstract
From 2010.igem.org
m (→How to solve Sudoku?) |
(→What is Sudoku?) |
||
| (47 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
= '''Sudoku''' = | = '''Sudoku''' = | ||
| + | <html> | ||
| + | <div> | ||
| + | <ul id="inpagemenu"> | ||
| + | <li><span>Introduction</span></li> | ||
| + | <li><a href="/Team:UT-Tokyo/Sudoku_construct" id="construct">System</a></li> | ||
| + | <li><a href="/Team:UT-Tokyo/Sudoku_modeling" id="modeling">Modeling</a></li> | ||
| + | <li><a href="/Team:UT-Tokyo/Sudoku_experiments" id="experiment">Experiments</a></li> | ||
| + | <li><a href="/Team:UT-Tokyo/Sudoku_perspective" id="perspective">Perspective</a></li> | ||
| + | <li><a href="/Team:UT-Tokyo/Sudoku_reference" id="reference">Reference</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <div id="clear"></div> | ||
| + | </html> | ||
| - | + | == '''Introduction''' == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | == ''' | + | ==='''Background'''=== |
| + | The ability to integrate and process information is indispensable for biological devices that operate autonomously in intricate environments, such as in "smart drugs", which sense and respond to the patient's health state appropriately. | ||
| + | Therefore, the design and construction of such genetic circuits has long been pursued in the field of synthetic biology since its inauguration. Accomplishments include the toggle switch, logic gate, counter, and flip-flop switch. | ||
| - | + | However, the function of most of these circuits is either limited to relatively simple form, such as Boolean logic element, or not robust enough to work correctly. A universal means of communication between individual elements is also generally lacking. As part of the solution, here we propose a new information processing unit which receives multiple inputs, memorizes the inputs stably, and makes a decision based on the combination of the inputs. In addition, we have devised a means of communication between individual bacterial-based elements. Importantly, this system allows destination-specific delivery of information even when the number of possible destinations is immensely large. As proof-on-principle of the above systems, we have created bacteria that solve "Sudoku", a world famous puzzle game. | |
==='''What is Sudoku?'''=== | ==='''What is Sudoku?'''=== | ||
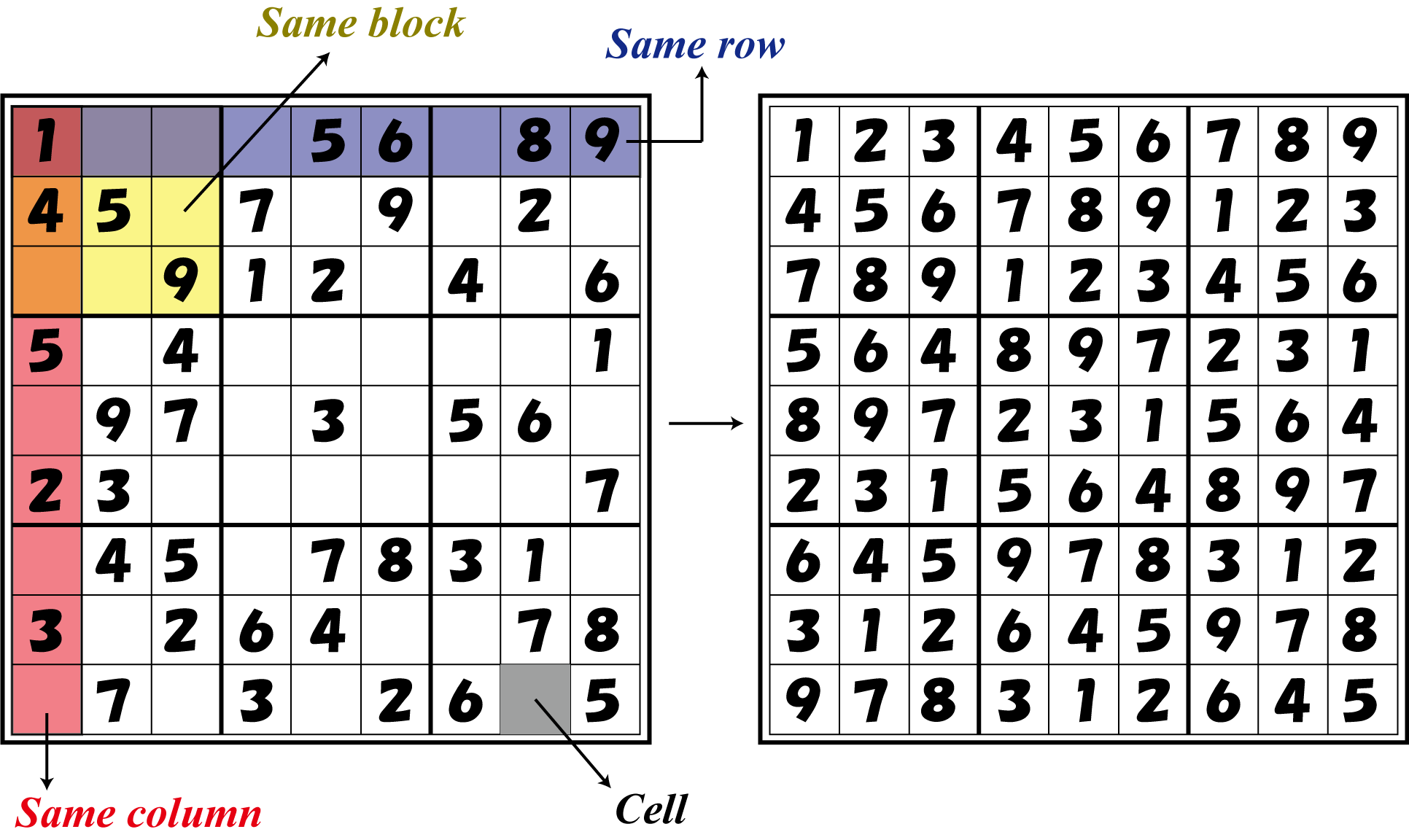
| - | [[Image: | + | [[Image:9x9.png|200px|thumb|The example of Sudoku (9x9)]] |
| - | Sudoku is a puzzle game with the objective of filling a 9x9 grid of cells with the numbers 1~9 without entering the same number in a column, row or “block (see figure).” | + | Sudoku is a puzzle game with the objective of filling a 9x9 grid of cells with the numbers 1~9 without entering the same number in a column, row or “block (see figure).” A player is given a grid in which some of the cells are filled in from the beginning and must complete filling in the grid by entering the remaining numbers. The rules are simple, but some puzzles can get very difficult, and it attracts fans from all over the world. For simplicity, here we solve a 4x4 grid version. However, expanding on the same principles, our ''E. coli'' can theoretically solve larger grids, for example 9x9 grids. |
| - | A player is given a grid in which some of the cells are filled in from the beginning and must complete filling in the grid by entering the remaining numbers. | + | |
| + | ==='''Solving Sudoku with our ''E. coli'''''=== | ||
| + | [[Image:E.coli corresponding.png|200px|thumb|16 kinds of ''E. coli'' corresponding to each cell]] | ||
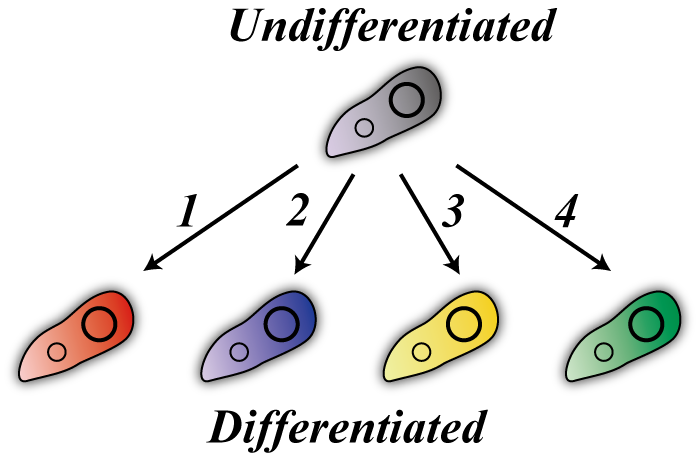
| - | + | [[Image:Differentiation-model.png|200px|thumb|Every cells can "differentiate"]] | |
| - | [[Image: | + | |
| - | + | Now, we explain how to make ''E. coli'' solve Sudoku. First, we make 16 kinds of ''E. coli'' corresponding to each cell. | |
| + | Our E. coli are in one of two states, which we designate “differentiated” or “undifferentiated.” There are four differentiated states corresponding to the numbers from one to four. | ||
| - | + | Differentiated ''E. coli'' have the ability to inform other bacteria what number they are, so that relevant bacteria do not differentiate into that number. Some ''E. coli'' are differentiated from the beginning. These bacteria set in action a chain of transmission events that result in the differentiation of bacteria corresponding to all 16 cells. | |
| - | + | These events take place in a flask which contains a co-culture of the 16 types of bacteria. Each of these 16 types interacts with detection bacteria on a plate which in turn inform the viewer the number which the 16 cells have differentiated into with the use of fluorescent proteins. | |
| - | + | ||
| - | + | If you'd like to know how our ''E.coli'' solves SUDOKU particularly, please see the [https://2010.igem.org/Team:UT-Tokyo/Sudoku_construct System] page. | |
| - | |||
| - | + | <html> | |
| - | + | <object width="640" height="385"><param name="movie" value="http://www.youtube.com/v/wzcryKryME8?fs=1&hl=ja_JP"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/wzcryKryME8?fs=1&hl=ja_JP" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="640" height="385"></embed></object> | |
| + | </html> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{{UT-Tokyo_Foot}} | {{UT-Tokyo_Foot}} | ||
Latest revision as of 03:57, 28 October 2010


Sudoku
Introduction
Background
The ability to integrate and process information is indispensable for biological devices that operate autonomously in intricate environments, such as in "smart drugs", which sense and respond to the patient's health state appropriately. Therefore, the design and construction of such genetic circuits has long been pursued in the field of synthetic biology since its inauguration. Accomplishments include the toggle switch, logic gate, counter, and flip-flop switch.
However, the function of most of these circuits is either limited to relatively simple form, such as Boolean logic element, or not robust enough to work correctly. A universal means of communication between individual elements is also generally lacking. As part of the solution, here we propose a new information processing unit which receives multiple inputs, memorizes the inputs stably, and makes a decision based on the combination of the inputs. In addition, we have devised a means of communication between individual bacterial-based elements. Importantly, this system allows destination-specific delivery of information even when the number of possible destinations is immensely large. As proof-on-principle of the above systems, we have created bacteria that solve "Sudoku", a world famous puzzle game.
What is Sudoku?
Sudoku is a puzzle game with the objective of filling a 9x9 grid of cells with the numbers 1~9 without entering the same number in a column, row or “block (see figure).” A player is given a grid in which some of the cells are filled in from the beginning and must complete filling in the grid by entering the remaining numbers. The rules are simple, but some puzzles can get very difficult, and it attracts fans from all over the world. For simplicity, here we solve a 4x4 grid version. However, expanding on the same principles, our E. coli can theoretically solve larger grids, for example 9x9 grids.
Solving Sudoku with our E. coli
Now, we explain how to make E. coli solve Sudoku. First, we make 16 kinds of E. coli corresponding to each cell.
Our E. coli are in one of two states, which we designate “differentiated” or “undifferentiated.” There are four differentiated states corresponding to the numbers from one to four.
Differentiated E. coli have the ability to inform other bacteria what number they are, so that relevant bacteria do not differentiate into that number. Some E. coli are differentiated from the beginning. These bacteria set in action a chain of transmission events that result in the differentiation of bacteria corresponding to all 16 cells. These events take place in a flask which contains a co-culture of the 16 types of bacteria. Each of these 16 types interacts with detection bacteria on a plate which in turn inform the viewer the number which the 16 cells have differentiated into with the use of fluorescent proteins.
If you'd like to know how our E.coli solves SUDOKU particularly, please see the System page.
 "
"