Team:UT-Tokyo/Sudoku abstract
From 2010.igem.org
m (→Solving Sudoku with our E. coli) |
|||
| Line 20: | Line 20: | ||
==='''Solving Sudoku with our E. coli'''=== | ==='''Solving Sudoku with our E. coli'''=== | ||
| - | [[Image: | + | [[Image:Differenciation-model.png|200px|thumb|Every cells can "differentiate"]] |
Now, we explain how to make E. coli solve Sudoku. First, we make 16 kinds E. coli corresponding to each cell. | Now, we explain how to make E. coli solve Sudoku. First, we make 16 kinds E. coli corresponding to each cell. | ||
(図!) | (図!) | ||
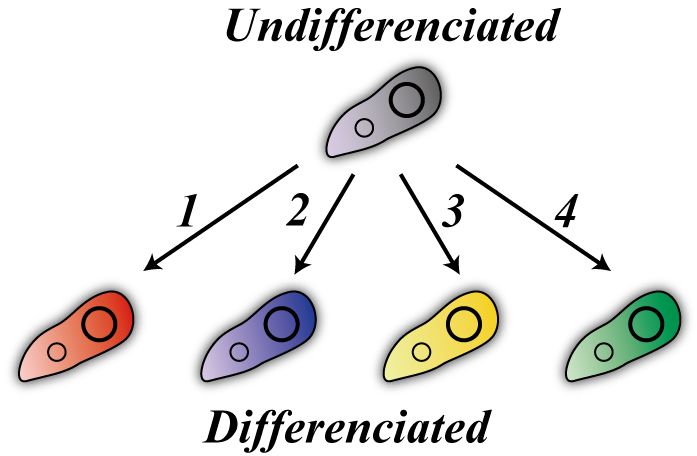
Our E. coli are in one of two states, which we designate “differentiated” or “undifferentiated.” There are four differentiated states corresponding to the numbers one to four. | Our E. coli are in one of two states, which we designate “differentiated” or “undifferentiated.” There are four differentiated states corresponding to the numbers one to four. | ||
| - | + | ||
Differentiated E. coli have the ability to inform other bacteria what number they are, so that relevant bacteria do not differentiate into that number. Some E. coli are differentiated from the beginning. These bacteria set in action a chain of transmission events that result in the differentiation of bacteria corresponding to all 16 cells. | Differentiated E. coli have the ability to inform other bacteria what number they are, so that relevant bacteria do not differentiate into that number. Some E. coli are differentiated from the beginning. These bacteria set in action a chain of transmission events that result in the differentiation of bacteria corresponding to all 16 cells. | ||
These events take place in a flask which contains a co-culture of the 16 types of bacteria. Each of these 16 types interacts with detection bacteria on a plate which in turn inform the viewer the number the 16 cells have differentiated into with the use of fluorescent proteins. | These events take place in a flask which contains a co-culture of the 16 types of bacteria. Each of these 16 types interacts with detection bacteria on a plate which in turn inform the viewer the number the 16 cells have differentiated into with the use of fluorescent proteins. | ||
Revision as of 22:59, 26 October 2010


Sudoku
Introduction System Lab note Result
Introduction
We're trying to make E.coli solve Sudoku puzzle. Human and Computers can solve Sudoku, of course. But E.coli, which is lower animal, solves sudoku in our project. It is very very interesting!
What is Sudoku?
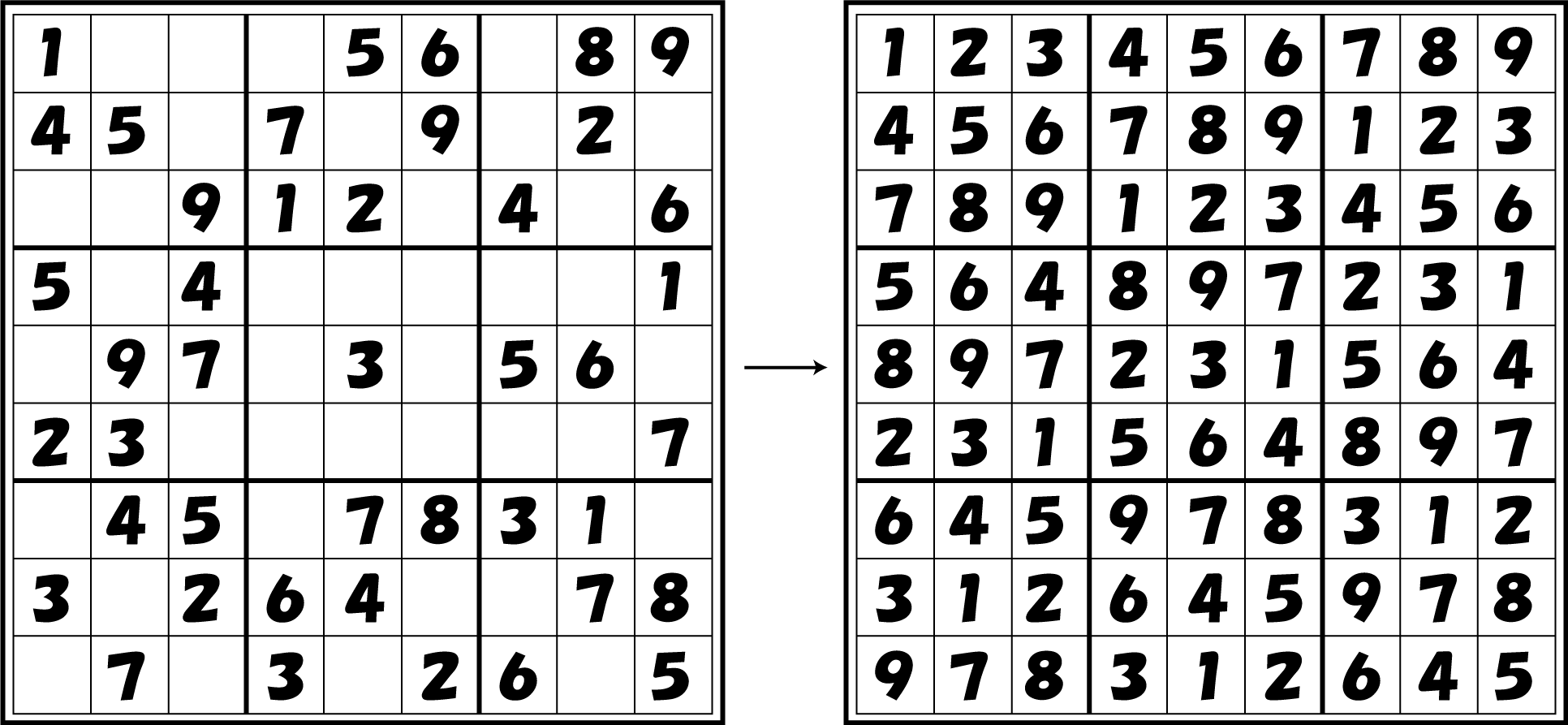
Sudoku is a puzzle game with the objective of filling a 9x9 grid of cells with the numbers 1~9 without entering the same number in a column, row or “block (see figure).” A player is given a grid in which some of the cells are filled in from the beginning and must complete filling in the grid by entering the remaining numbers. The rules are simple, but some puzzles can get very difficult, and it attracts fans from all over the world. For simplicity, here we solve a 4x4 grid version. However, expanding on the same principles, our E. coli can theoretically solve larger grids, for example 9x9 grids.
Solving Sudoku with our E. coli
Now, we explain how to make E. coli solve Sudoku. First, we make 16 kinds E. coli corresponding to each cell. (図!) Our E. coli are in one of two states, which we designate “differentiated” or “undifferentiated.” There are four differentiated states corresponding to the numbers one to four.
Differentiated E. coli have the ability to inform other bacteria what number they are, so that relevant bacteria do not differentiate into that number. Some E. coli are differentiated from the beginning. These bacteria set in action a chain of transmission events that result in the differentiation of bacteria corresponding to all 16 cells. These events take place in a flask which contains a co-culture of the 16 types of bacteria. Each of these 16 types interacts with detection bacteria on a plate which in turn inform the viewer the number the 16 cells have differentiated into with the use of fluorescent proteins.
Reference
as RNA
1.Artificial antisense RNAs silence lacZ in E. coli by decreasing target mRNA concentration Stefan Alessandra, Tonelli Alessandro, Schwarz Flavio and Hochkoeppler Alejandro, BMB reports, 568-74 (2008)
2.Kill the messenger: bacterial antisense RNA promotes mRNA decay E Gerhart H Wagner, nature structural & molecular biology, 16, 804-6 (2009)
3.Distance between RBS and AUG plays an important role in overexpression of recombinant proteins Sunil K. Berwal, R.K. Sreejith and Jayanta K. Pal, Analytical Biochemistry, 405, 275-7 (2010)
4.Antisense RNA Control of Plasmid R1 Replication Charlotta Malmgren, E. Gerhart H. Wagner, Chantal Ehresmann, Bernard Ehresmann and Pascale Romby, The Journal of Biologycal Chemistry, 272, 12508-12 (1997)
5.Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli Nobutaka Nakashima, Tomohiro Tamura and Liam Good, Nucleic Acids Research, 34, e138 (2006)
6.The Influence of Antisense Oligonucleotide-induced RNA Structure on Escherichia coli RNase H1 Activity Walt F. Lima, Venkatraman Mohan and Stanley T. Crooke, The Journal of Biological Chemistry, 272, 18191-8 (2010)
MS2 phage
1.Studies on Sex Pili: Mutants of the Sex Factor F in Escherichia coli Defective in Bacteriophage-Adsorbing Function of F Pili Munemitsu Tomoeda, Akemi Shuta and Manabu Inuzuka, Journal of Bacteriology, 112, 1358-63 (1972)
2.Crystal structure of an RNA bacteriophage coat protein-operator complex Karin Valegard, James B. Murray, Peter G. Stockley, Nicola J. Stonehouse and Lars Liljas, Nature, 371, 626 (1994)
3.Ribonucleoprotein Complexes of R17 Coat Protein and a Translational Operator Analog Dorothy Beckett and Olke C. Uhlenbeck, Journal of Molecular Biology, 204, 927-38 (1988)
4.A unique mechanism regulating gene expression: Translational inhibition by a complementary RNA transcript (micRNA) Takeshi Mizuno, Mei-Yin Chou and Masayori Inoue, Proc. Nail. Acad. Sci. USA, 81, 1966-70 (1984)
The others
1.Purnick, P.E., and Weiss, R. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell. Biol. 10, 410?422.(2009).
2.Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339?342 (2000).
3.Anderson, J. C., Clarke, E. J., Arkin, A. P. & Voigt, C. A. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 355, 619?627 (2006).
4.Anderson, J.C., Voigt, C.A. & Arkin, A.P. Environmental signal integration by a modular AND gate. Mol. Syst. Biol. 3, 133 (2007).
5.Win, M. N. & Smolke, C. D. Higher-order cellular information processing with synthetic RNA devices. Science 322, 456?460 (2008).
6.Ham, T. S., Lee, S. K., Keasling, J. D. & Arkin, A. P. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS ONE 3, e2815 (2008).
7.Friedland, A. et al. Synthetic gene networks that count. Science 324, 1199?1202 (2009).
 "
"