|
JULY: WEEK 5
July, 26th
Ligation of I15_4C5=I15(E-P)+4C5(E-P)
We retrieved from our freezer I15-1 (quantified 34,2 ng/ul) and 4C5 (quantified 24,0 ng/ul)
Digestion E-P was performed:
| Culture | Kind | Final reaction volume (ul) | DNA (ul) | H20 (ul) | Enzyme 1 | Enzyme 2 | Buffer H
|
| I15-1 | Insert | 25 | 18,5 | 2 | 1 EcoRI | 1 PstI | 2,5
|
| pSB4C5 | Vector | 25 | 3,6 | 16,9 | 1 EcoRI | 1 PstI | 2,5
|
Digestions were incubated at 37°C for 3 hours. A medium 1% agarose gel was prepared. I15-1 didn't give good results, in fact many unwanted extra-bands were observed. For this reason, we decided not to perform ligation, but to sequence I15-1. An inoculum from glycerol stock for I15-1 was performed in 1ml LB+Amp, and incubated at 37°C 220rpm ON. Tomorrow I15-1 plasmide will be extracted with MiniPrep kit and I15-1 sample will be prepared for sequencing.
Today we also screened 3 contaminants of MC1061 transformed with RING, incubated yesterday
 4C5 (E-P) and I15-1 (E-P): extra-bands can be observed for I15-1 |
 MC1-2-3 contaminants screening: no plasmid was observed |
July, 27th
Today we prepared I15-1 sample for sequencing. MiniPrep was performed on this culture and samples for both forward and reverse sequencing were prepared and sent to BMR genomics.
Our self-inducible parts assembly goes on: we are ready to co-trasform I14_4C5, I16_4C5, I17_4C5, I18_4C5 and I19_4C5 in T9002 competent strain (home made).
Since we noticed that our I7-3 and I8-5 glycerol stocks are not present in our freezer, we decided to prepare them today, starting from purified DNA (from MiniPrep) and tranforming it again in TOP10.
| Ligation name | E. coli strain | Resistance |
- I14_4C5-2= I14 (E-P) + pSB4C5 (E-P)
|
T9002
|
Amp 100+Cm 12,5 |
- I16_4C5-1= I16 (E-P) + pSB4C5 (E-P)
|
T9002
|
Amp 100+Cm 12,5 |
- I17_4C5-1= I17 (E-P) + pSB4C5 (E-P)
|
T9002
|
Amp 100 + Cm 12,5 |
- I18_4C5-1= I18 (E-P) + pSB4C5 (E-P)
|
T9002
|
Amp 100 + Cm 12,5 |
- I19_4C5-1= I19 (E-P) + pSB4C5 (E-P)
|
T9002
|
Amp 100 + Cm 12,5 |
|
|
TOP10
|
Amp 100 |
|
|
TOP10
|
Amp 100 |
MiniPrep was performed also on MC1 and MC2 (colonies grown on LB+Cm 12,5 for MC1061)
- MC1: 129,8 ng/ul, digested with HindIII
- MC2: 34 ng/ul, digested with HindIII
 Screening for MC1 and MC2: digested with HindIII and undigested July, 28th
Today we received primers to modify PhaPs. We diluted primers to perform a PCR with them in order to mutagenize this BioBrick to get its prefix Standard/Silver compliant and suffix Silver compliant.
July, 29th
Today we performed PCR in order to amplify the phasin PhaP (<partinfo>BBa_K208001</partinfo>). Out primers are built to eliminate the stop codon from phasin and to give it the prefix and suffix we desire:
We used our new synthesized primers to modify through PCR <partinfo>BBa_K208001</partinfo> phasin in order to create two new parts without stop codon but with Standard prefix and Silver suffix the first one, Silver prefix and suffix the second one.
 Gel run for PCR-modified phasins Gel extraction for phasins showed the following quantifications:
- Pha-10S-1: 15 ng/ul
- Pha-10S-2: 18,5 ng/ul
- Pha-SS-1: 22,8 ng/ul
- Pha-SS-2: 22,4 ng/ul
Phasins were digested X-S for 3 hours and were quantified:
- Pha-10S-1 (X-S): 14 ng/ul
- Pha-10S-2 (X-S): 13,2 ng/ul
- Pha-SS-1 (X-S): 15 ng/ul
- Pha-SS-2 (X-S): 14,9 ng/ul
Competentization of MC1061 (again) because we suspect the presence of a contaminant (it grew without reason on Cm plates).
Transformation of new MC1061 competent cells with:
- 1 ul (4ng) of miniprepped ENTERO-pSB4C5 (positive control);
- 1 ul of RING ligation (RING shouldn't be propagated in MC1061);
- 1 ul of MilliQ (negative control).
Transformed cells have been plated on Cm 12,5 ug/ml agar plates and incubated overnight at 37°C.
Tecan Test
July, 30th
We checked the presence of colonies in plates.
 MC1061 transformed with ENTERO-pSB4C5 (positive control) |  MC1061 transformed with RING |  MC1061 transformed with MilliQ (negative control) |
We calculated (thanks Federica) efficiency as #colonies/ug DNA plated
| Strain | Vector | #colonies | Efficiency
|
| MC1061 | ENTERO-pSB4C5 | 3700 | ~10^6
|
| MC1061 | RING | 0 | -
|
| MC1061 | NOTHING | 0 | -
|
As espected RING and NOTHING didn't grow.
MC1061 competent cells don't replicate RING :) !!!
Miniprep of <partinfo>J04450</partinfo> to take the vector <partinfo>pSB1A3</partinfo>.
3-hour digestion (X-S) and gel run to extract the backbone (~2157 bp).
| Culture | Kind | Final reaction volume (ul) | DNA (ul) | H20 (ul) | Enzyme 1 (ul) | Enzyme 2 (ul) | Buffer H
|
| <partinfo>BBa_J04450</partinfo> | Vector | 25 | 17,2 | 3,3 | 1 X | 1 S | 2,5
|
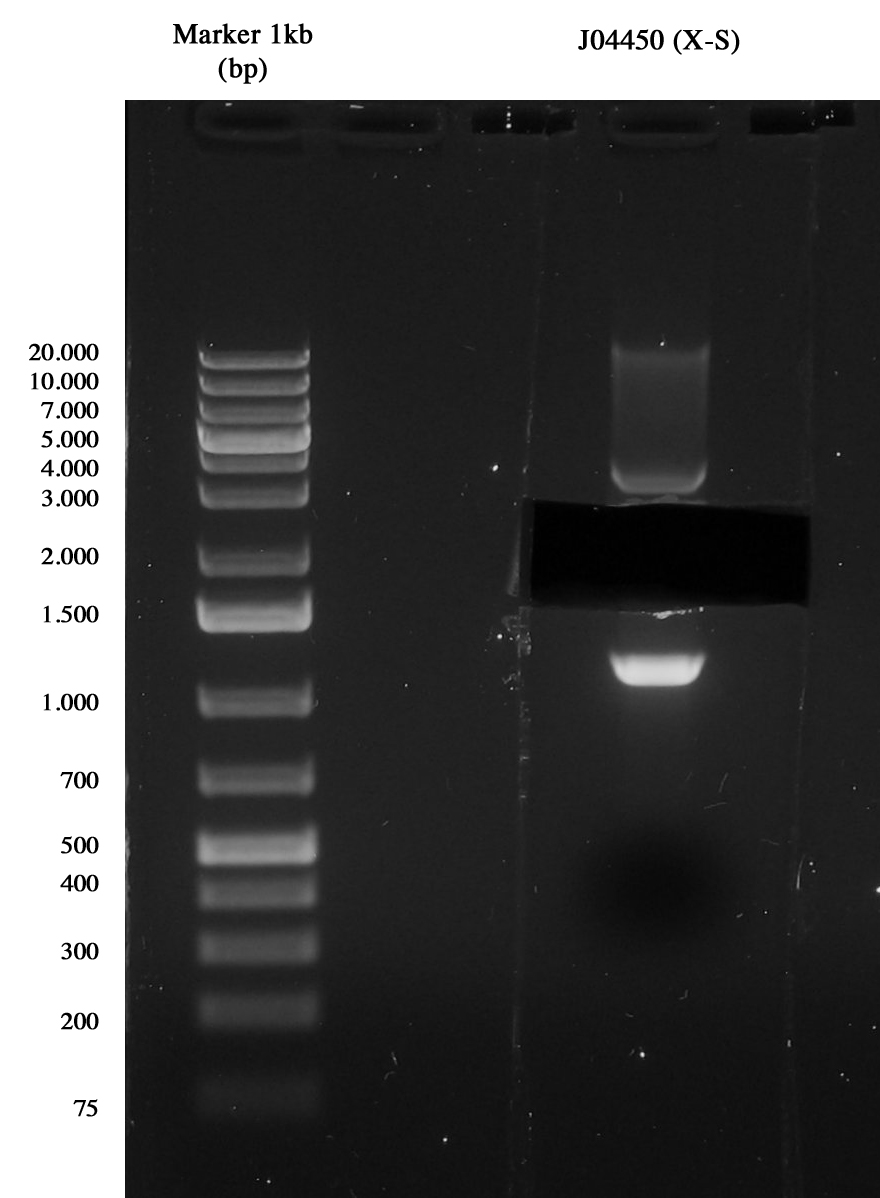 Gel run of J04450 to take pSB1A3 vector Gel extraction was quantified 15,5 ng/ul.
Ligation of:
- I20: Pha-10S-1 (X-S) + pSB1A3 (X-S)
- I21: Pha-SS-1 (X-S) + pSB1A3 (X-S)
July, 31st
Transformation of I20 and I21 into E. coli DH5-alpha. Cells were plated on LB+Amp agar plates and grown overnight at 37°C.
August, 1st
All plates showed colonies (there were also red colonies that contained a wrong ligated plasmid). So we picked three colonies each plate (the right ones) and inoculated them into 5 ml LB+Amp. They were let grow ON at 37°C, 220 rpm.
|
|
 "
"










