Team:UNIPV-Pavia/Calendar/August/settimana1
From 2010.igem.org
m (→August, 5th) |
|||
| (24 intermediate revisions not shown) | |||
| Line 30: | Line 30: | ||
<html><p align="center"><font size="4"><b>AUGUST: WEEK 1</b></font></p></html><hr><br> | <html><p align="center"><font size="4"><b>AUGUST: WEEK 1</b></font></p></html><hr><br> | ||
| + | <html><a name="indice"/></html> | ||
==August, 2nd== | ==August, 2nd== | ||
Miniprep and quantification with Nanodrop of: | Miniprep and quantification with Nanodrop of: | ||
| Line 108: | Line 109: | ||
Gel run of amplified DNA showed for every sample the expected distance between primers of a strain with nothing integrated in attPhi80 site (~570 bp), but unfortunately we forgot to take a picture of the gel ;( | Gel run of amplified DNA showed for every sample the expected distance between primers of a strain with nothing integrated in attPhi80 site (~570 bp), but unfortunately we forgot to take a picture of the gel ;( | ||
| + | |||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==August, 4th== | ==August, 4th== | ||
| Line 131: | Line 135: | ||
[[Image:UNIPV10_plux.jpg|thumb|200px|center|Autoinducibles]] | [[Image:UNIPV10_plux.jpg|thumb|200px|center|Autoinducibles]] | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
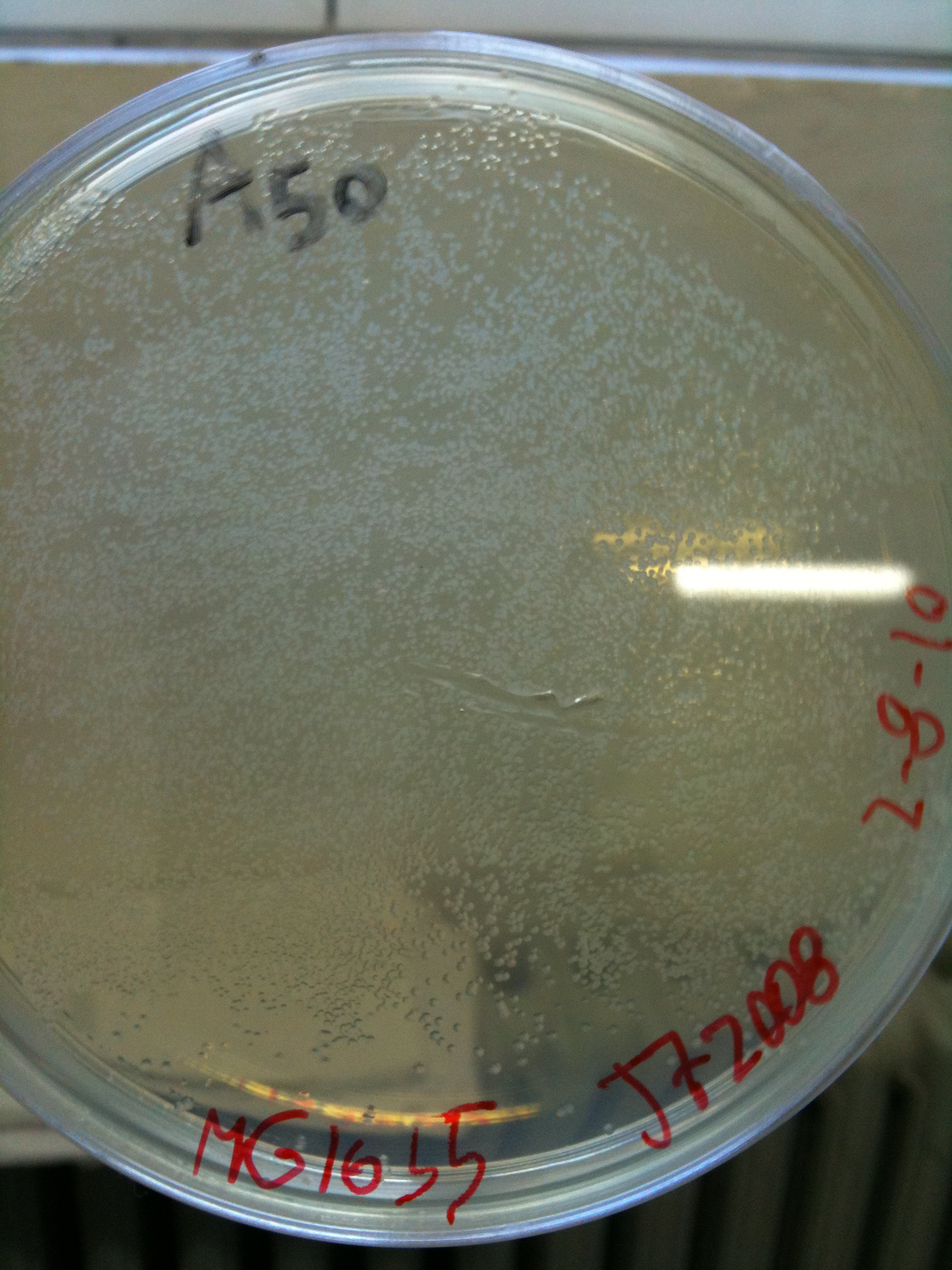
==August, 5th== | ==August, 5th== | ||
| - | + | Glycerol stock and miniprep of MG1655 and MC1061 cultures incubated for 19 hours at 30°C, 220 rpm. | |
Miniprep was quantified as follows: | Miniprep was quantified as follows: | ||
*MG123-1: 27 ng/ul | *MG123-1: 27 ng/ul | ||
| Line 150: | Line 156: | ||
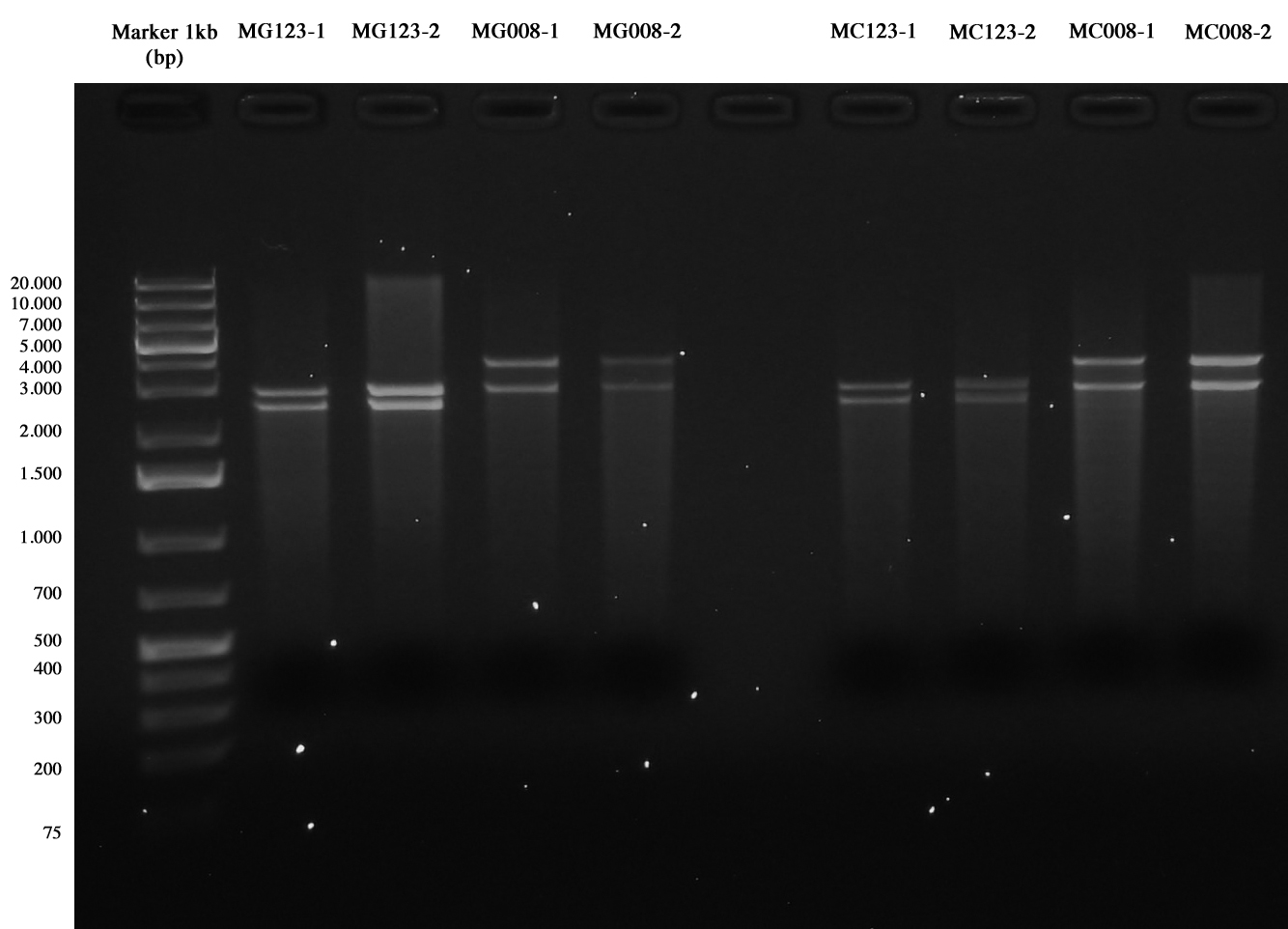
[[Image:UNIPV10_transformed_helper_screening.jpg|thumb|200px|center|pAH123 and <partinfo>BBa_J72008</partinfo> screening (miniprepped from MG1655 and MC1061)]] | [[Image:UNIPV10_transformed_helper_screening.jpg|thumb|200px|center|pAH123 and <partinfo>BBa_J72008</partinfo> screening (miniprepped from MG1655 and MC1061)]] | ||
| - | + | This time gel run succeeded, we chose samples | |
| + | *MG123-1 | ||
| + | *MG008-1 | ||
| + | *MC123-1 | ||
| + | *MC008-1 | ||
| + | to be re-competentizied. So they were inoculated into 5 ml LB+Amp 50 ug/ml and grown and shaken ON at 30°C. | ||
---- | ---- | ||
| - | + | Sequencing for I20 and I21 arrived from BMR, but all samples were wrong; sites X and S of the vector paired and nothing could ligate. So we started a new ligation cycle dephosphorylating the previously gel-extracted vector <partinfo>pSB1A3</partinfo>. | |
| + | New ligations: | ||
| + | *I20-new: Pha-10S-1 (X-S) + <partinfo>pSB1A3</partinfo> (X-S, dephosphorylated) | ||
| + | *I21-new: Pha-SS-1 (X-S) + <partinfo>pSB1A3</partinfo> (X-S, dephosphorylated) | ||
| + | and their negative control: | ||
| + | *C-: <partinfo>pSB1A3</partinfo> (X-S, dephosphorylated) | ||
| + | |||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==August, 6th== | ==August, 6th== | ||
| + | Resuspension of linker <partinfo>BBa_K105012</partinfo> from iGEM 2010 Distribution Kit. | ||
| + | |||
| + | Transformation of ligations and resuspended DNA | ||
| + | *I20-new | ||
| + | *I21-new | ||
| + | *C- | ||
| + | *<partinfo>BBa_K105012</partinfo> | ||
| + | into 100ul ''E. coli DH5-alpha''. | ||
| + | |||
| + | They were plated on LB+Amp 100ug/ml agar plates | ||
| + | |||
| + | ---- | ||
| + | Competentization of colonies selected the previous day: | ||
| + | *MG123 (without '-1' from now on) | ||
| + | *MG008 | ||
| + | *MC123 | ||
| + | *MC008 | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==August, 7th== | ==August, 7th== | ||
| + | Plates of transformed cells were stored at +4°C. | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| - | |||
| + | <!-- table previous next week --> | ||
<br><br> | <br><br> | ||
<table border="0" width="100%" class="menu"> | <table border="0" width="100%" class="menu"> | ||
| Line 168: | Line 209: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | <!-- fine table previous next week --> | ||
</td> | </td> | ||
Latest revision as of 16:58, 24 October 2010
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"