Team:UNAM-Genomics Mexico/Modeling
From 2010.igem.org
| Line 28: | Line 28: | ||
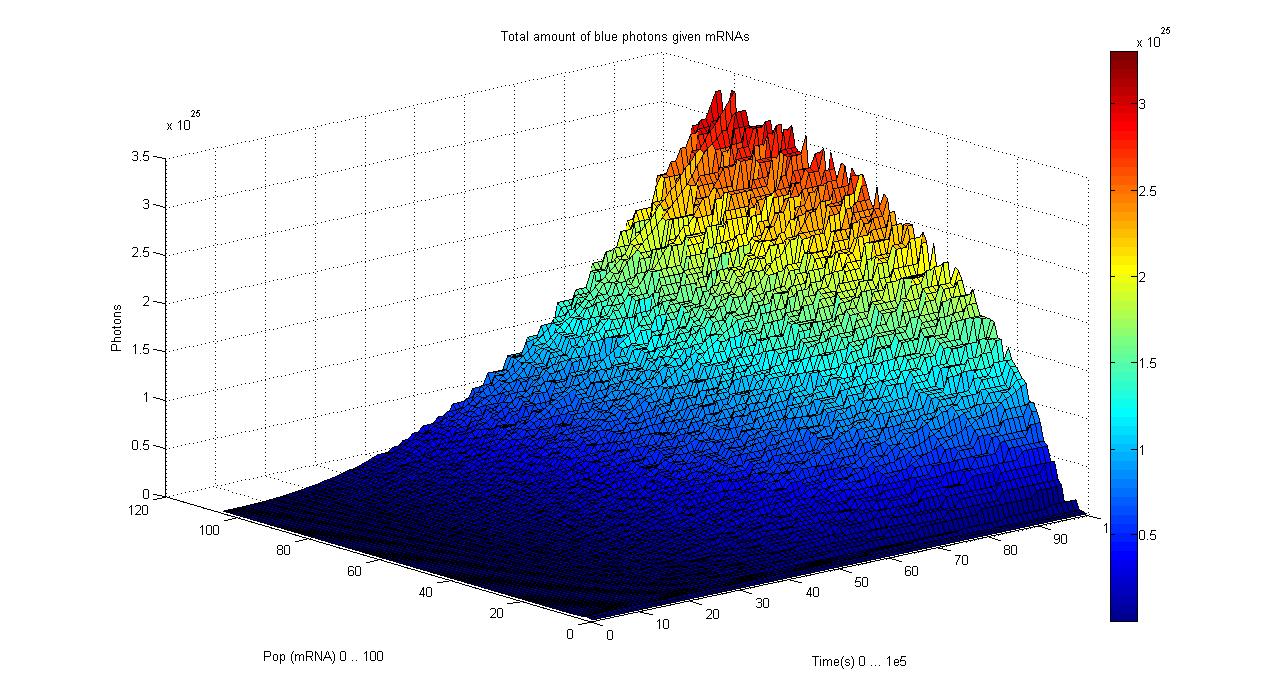
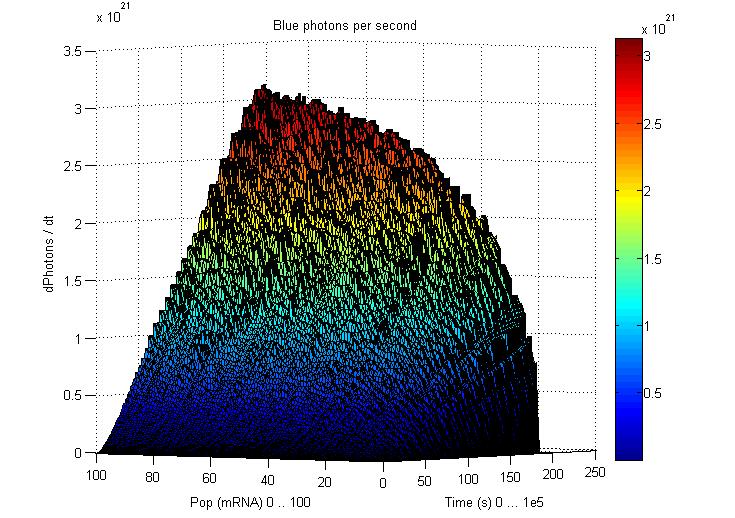
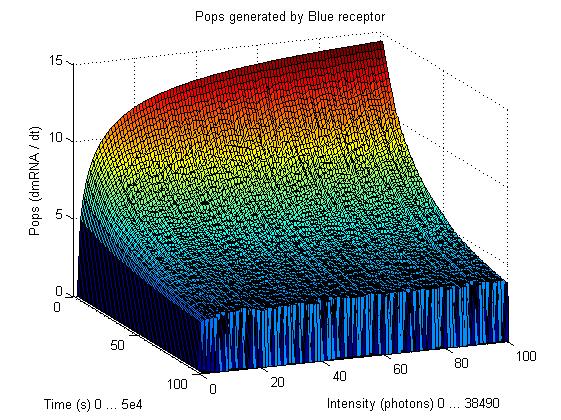
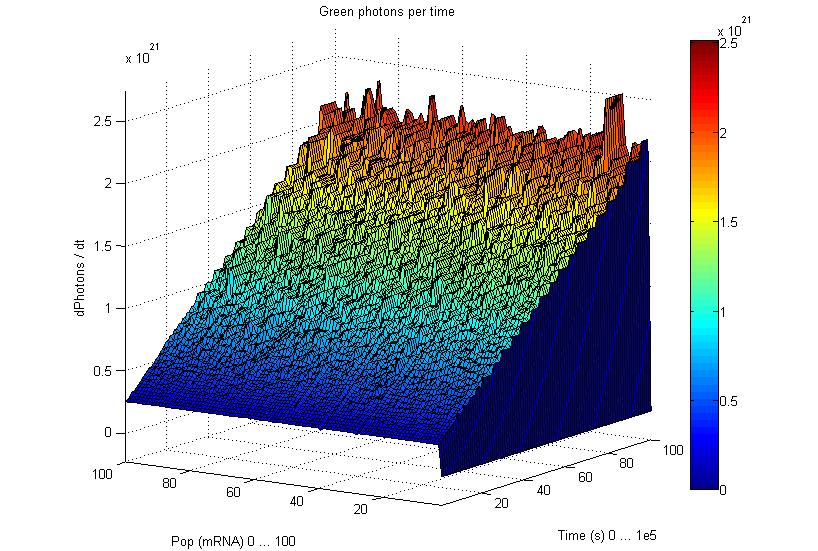
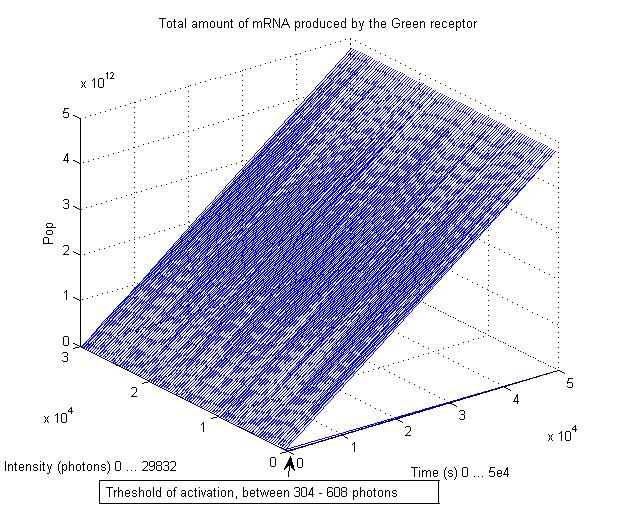
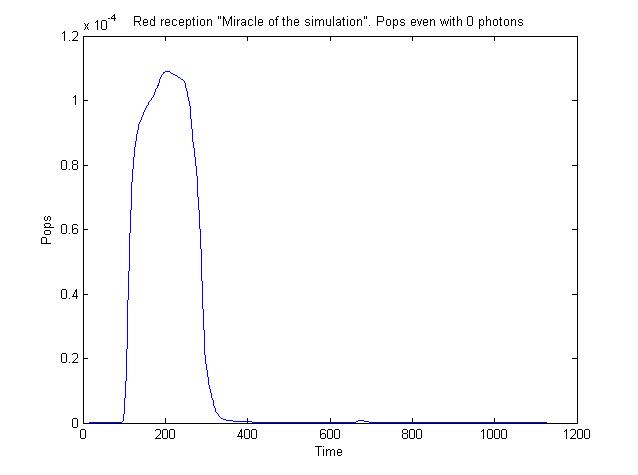
So having Simbiolgy, we decided to model our interfaces using it's deterministic solvers. Some results are here: | So having Simbiolgy, we decided to model our interfaces using it's deterministic solvers. Some results are here: | ||
| - | + | ||
| - | image:UNAM-Genomics_Mexico_Blue_emission_phot.jpg | + | [[image:UNAM-Genomics_Mexico_Blue_emission_phot.jpg]] |
| - | image:UNAM-Genomics_Mexico_Blue_emission_phots.jpg | + | |
| - | image:UNAM-Genomics_Mexico_Blue_reception_Pops.jpg | + | [[image:UNAM-Genomics_Mexico_Blue_emission_phots.jpg]] |
| - | image:UNAM-Genomics_Mexico_Green_emission_phots.jpg | + | |
| - | image:UNAM-Genomics_Mexico_Green_receptor_Pop.jpg | + | [[image:UNAM-Genomics_Mexico_Blue_reception_Pops.jpg]] |
| - | image:UNAM-Genomics_Mexico_Miracle.jpg | + | |
| - | + | [[image:UNAM-Genomics_Mexico_Green_emission_phots.jpg]] | |
| + | |||
| + | [[image:UNAM-Genomics_Mexico_Green_receptor_Pop.jpg]] | ||
| + | |||
| + | [[image:UNAM-Genomics_Mexico_Miracle.jpg]] | ||
| + | |||
We chose to simulate the flux of output given a determined input. So we have for a wide range of inputs, the resultant outputs in a time manner. | We chose to simulate the flux of output given a determined input. So we have for a wide range of inputs, the resultant outputs in a time manner. | ||
| Line 43: | Line 48: | ||
Also, having the Kappa binaries, we decided to model the system using Kappa as a stochastic modeling platform. Some preliminary results would be here if the Server didn't have an unexplainable aversion for .7z, .rar, or indigestion for .zip | Also, having the Kappa binaries, we decided to model the system using Kappa as a stochastic modeling platform. Some preliminary results would be here if the Server didn't have an unexplainable aversion for .7z, .rar, or indigestion for .zip | ||
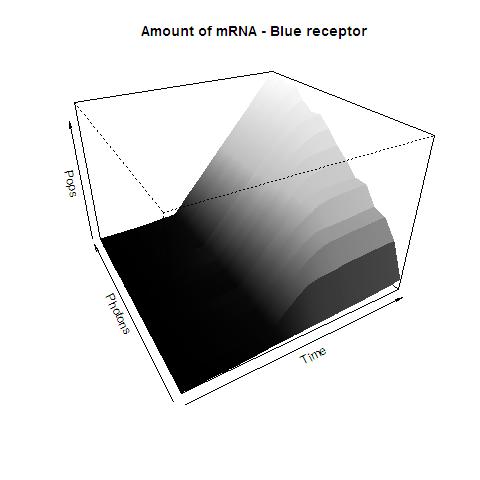
| - | As soon as the Server decides to tolerate our files, we will upload them. | + | As soon as the Server decides to tolerate our files, we will upload them. In the mean time, here's a sample graph. |
| + | [[image:UNAM-Genomics_Mexico_Blue_Receptor_Kappa.jpg]] | ||
| + | This graphs plots the PopS output of our Blue Receptor (lovTAP). The system begins in dark state, and at a given time (you can guess which one), there's a photon influx. This graph is however in arbitrary units. We wanted to make a more useful characterization, but these simulations required some extreme computing time. | ||
| + | Should we finish them, we will upload them. | ||
===Neural Network=== | ===Neural Network=== | ||
Revision as of 21:11, 26 October 2010

So, here goes the Model!
Simbiology
So having Simbiolgy, we decided to model our interfaces using it's deterministic solvers. Some results are here:
We chose to simulate the flux of output given a determined input. So we have for a wide range of inputs, the resultant outputs in a time manner.
Kappa
Also, having the Kappa binaries, we decided to model the system using Kappa as a stochastic modeling platform. Some preliminary results would be here if the Server didn't have an unexplainable aversion for .7z, .rar, or indigestion for .zip As soon as the Server decides to tolerate our files, we will upload them. In the mean time, here's a sample graph.
This graphs plots the PopS output of our Blue Receptor (lovTAP). The system begins in dark state, and at a given time (you can guess which one), there's a photon influx. This graph is however in arbitrary units. We wanted to make a more useful characterization, but these simulations required some extreme computing time. Should we finish them, we will upload them.
Neural Network
In Training... Having over a million data to map, our Levenberg-Marquandt algorithm is taking longer than expected.iGEM
iGEM is the International Genetically Engineered Machines Competition, held each year at MIT and organized with support of the Parts Registry. See more here.Synthetic Biology
This is defined as attempting to manipulate living objects as if they were man-made machines, specifically in terms of genetic engineering. See more here.Genomics
We are students on the Genomic Sciences program at the Center for Genomic Sciences of the National Autonomous University of Mexico, campus Morelos. See more here.This site is best viewed with a Webkit based Browser (eg: Google's Chrome, Apple's Safari),
Trident based (Microsoft's Internet Explorer) or Presto based (Opera) are not currently supported. Sorry.
 "
"