Team:TU Delft/Project/tolerance/parts
From 2010.igem.org

Survival Parts
Introduction to Solvent tolerance
The effect of organic solvents is widely known; mostly they can accumulate in the cell membrane changing the composition, thus affecting its physiological function. Our goal was to develop an E.coli strain capable of working in two-phase systems with resistance to high concentrations of hydrophobic molecules in the membrane.
Different papers were checked, some of them reporting an increase in the organic solvent tolerance of E.coli after the over-expression of efflux-pumps, ostA E.coli gene and some other genes. Efflux-pumps seemed to complex due to de amount of genes required and because of the fact that these systems also confer antibiotic resistance to the cells; on the other hand ostA gene is a common gene in E.coli, in order to have a negative control it is required to disrupt the gene which could make complex the characterization of our biobrick.
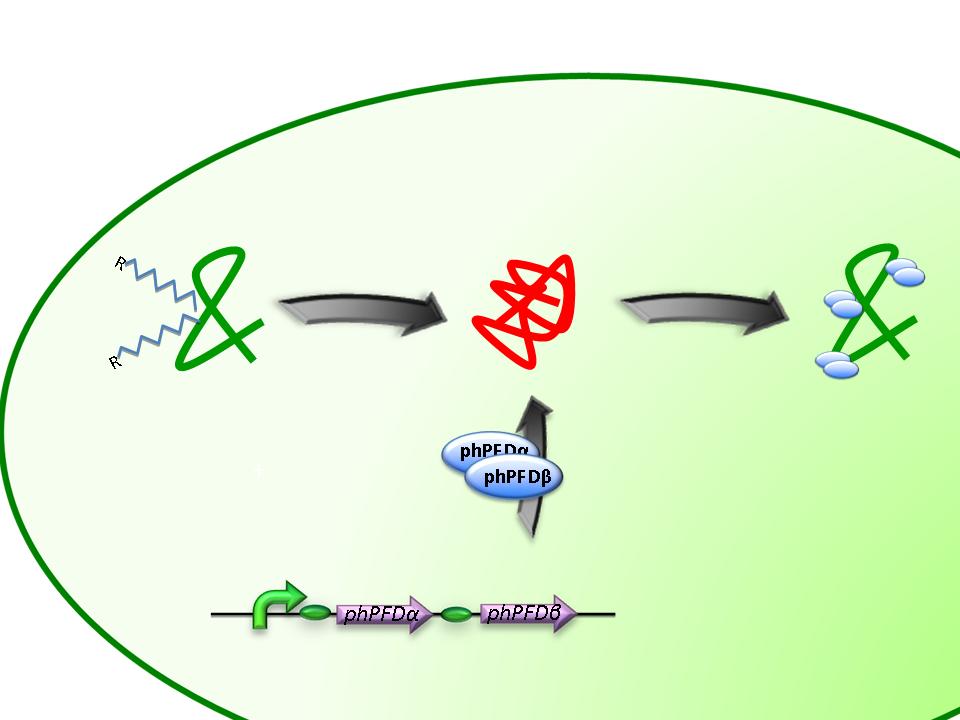
From the work of Okochi and co-workers, we discovered the prefoldin system from Pyrococcus horikoshii OT3. Prefoldin is a part of a widely spread group of proteins: chaperones. These proteins have the property of helping during the protein folding process, and are related also to thermal shock resistance. Therefore we opted for the use of two proteins; namely phPFDα and phPFDβ (Figure 1). These proteins increase the tolerance to alkanes by acting as chaperones, which correct protein misfolding /unfolding in the presence of alkanes.
BBa_K398406 - Prefoldin
J23100- phPFDα – J23100 phPFDβ View this part in the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398406 parts registry]
Introduction to Salt tolerance
Salts are essential for life, but not all life requires the same amount or the same type of salts. But because of these varying conditions the salt stress differs from environment to environment. It is difficult for a species to adapt to different salt stress levels. To compensate for the cells innate vulnerability to higher salt concentrations we chose a protein from Chlamydomonas sp. W80 that increases the salt tolerance of E.coli. This protein is called bbc1. The exact mechanism of the increased salt tolerance is as of yet unknown. But since bbc1 shows a high homology to RBS binding proteins, it is theorized that it may prevent/stabilize the folding structure of the ribosome under high salt stress conditions.
Genes for solvent and salt tolerance were found during our literature review: BBC1, prefoldin alpha and beta. The original sequences of these genes differ in composition from the E. coli sequences; furthermore they contain undesired restriction sites. We decided to enhance our sequences for a successful expression on E. coli, for that purpose we used jcat website tool.
Once we had the enhanced sequence, we added the standard Biobrick prefix and suffix. Finally, we sticked the promoter-RBS sequences that best suit for our CDS.
BBa_K39810 1/2/8 - bbc1
For salt tolerance multiple construct were made with the same protein, bbc1, but different expressions:
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398102 BBa_K398102]: J23100-bbc1-B0015, Low expression
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398101 BBa_K398101]: J23101-bbc1, Low to moderate expression
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398108 BBa_K398108]: R0040-B0034-bbc1-B0015, High expression
References
- S. Tanaka,K. Ikeda, H. Miyasaka, Enhanced Tolerance Against Salt-Stress and Freezing-Stress of Escherichia coli Cells Expressing Algal bbc1 Gene. Current Microbiology, 42:173-177 (2001)
- Y. Suda,T. Yoshikawa,Y. Okuda,M. Tsunemoto,S. Tanaka,K. Ikeda,H. Miyasaka,M. Watanabe,K. Sasaki,K. Harada,T. Bamba,K. Hirata, Isolation and characterization of a novel antistress gene from Chlamydomonas sp. W80. Journal of Bioscience and Bioengineering, 107(4) 352-354 (2009)
- Y. Hase, S. Yokoyama, A. Muto, et al. , Removal of a ribosome small subunit-dependent GTPase confers salt resistance on Escherichia coli cells. RNA Society, 15:1766-1774 (2009)
- Mihaela Marilena Lăzăroaie,Investigation of saturated and aromatichydrocarbon resistance mechanismsin Pseudomonas aeruginosa IBB Cent. Eur. J. Biol. 4(4) 469–481 (2009)
- M. Okochi, K. Kanie, M. Kurimoto ,M. Yohda & Hiroyuki Honda Overexpression of prefoldin from the hyperthermophilic archaeum Pyrococcus horikoshii OT3 endowed Escherichia coli with organic solvent tolerance Appl Microbiol Biotechnol 79:443–449 (2008)

 "
"