Team:TU Delft/Project/sensing/parts
From 2010.igem.org
| Line 19: | Line 19: | ||
| - | [[Image:TU Delft PalkS.PNG| | + | [[Image:TU Delft PalkS.PNG|450px|thumb|left|'''Figure 2''' – Schematic description of the alkane sensing pathway with the corresponding genes.]] |
| + | [[Image:TU_Delft_PCaif.PNG|450px|thumb|right|'''Figure 3''' – Schematic description of the alkane sensing pathway with the corresponding genes.]] | ||
'''Control by Crp''' | '''Control by Crp''' | ||
| Line 27: | Line 28: | ||
For the induction of alkane-degradation genes in this construct the BioBrick consisted out of the P(CaiF) promoter, AlkS gene and PalkB gene. (figure 3) | For the induction of alkane-degradation genes in this construct the BioBrick consisted out of the P(CaiF) promoter, AlkS gene and PalkB gene. (figure 3) | ||
| - | |||
===BBa_K398331: P(Caif)-RBS-GFP-TT=== | ===BBa_K398331: P(Caif)-RBS-GFP-TT=== | ||
Revision as of 20:38, 26 October 2010

Sensing Parts
Introduction
Catabolite repression control
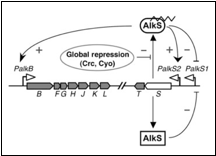
In Pseudomonas putida, the AlkS transcriptional regulator activates the expression of its own gene, and that of alkT, from a promoter named PalkS2 in the presence of alkanes. This allows achieving AlkS levels that are high enough to activate the expression of the alkBFGHJKL operon from the PalkB promoter. AlkS recognizes C5–C10 n-alkanes as effectors, but does not respond to shorter or larger alkanes (Rojo et al. 2009) (figure 1)
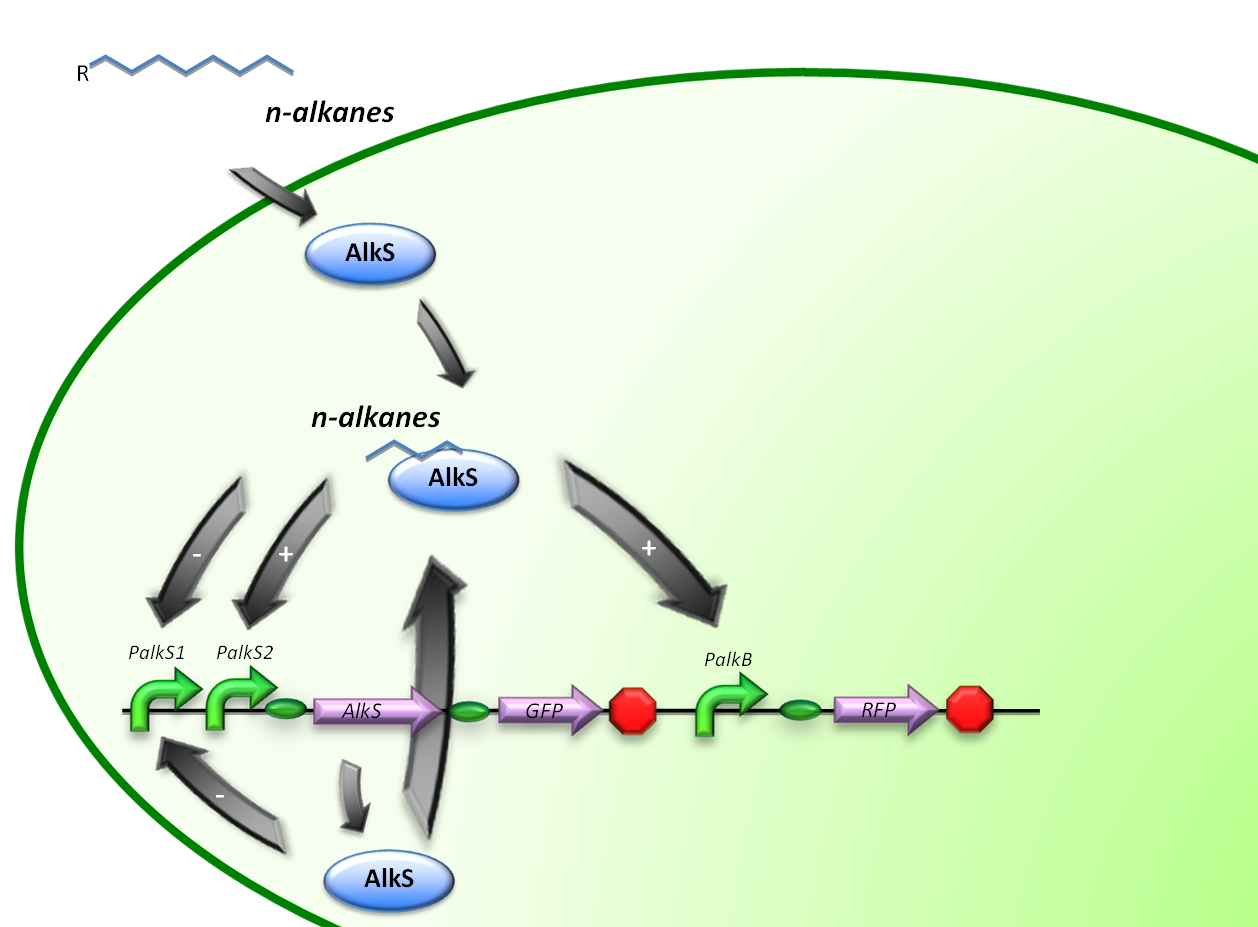
In stead of using all the alkBFGHJKL genes, a GFP will be introduced into our system to check the functionality of the PalkB promoter in E.coli. (figure 2)
Therefore, the first construct is suitable for the analysis of transcriptional activity over the PalkS promoter. A plasmid containing the AlkS-PalkS-PalkB regulatory mechanism will be coupled to GFP and RFP generators in order to determine transcriptional activities of PalkS and PalkB at varying hydrocarbon concentrations by measuring fluorescence.
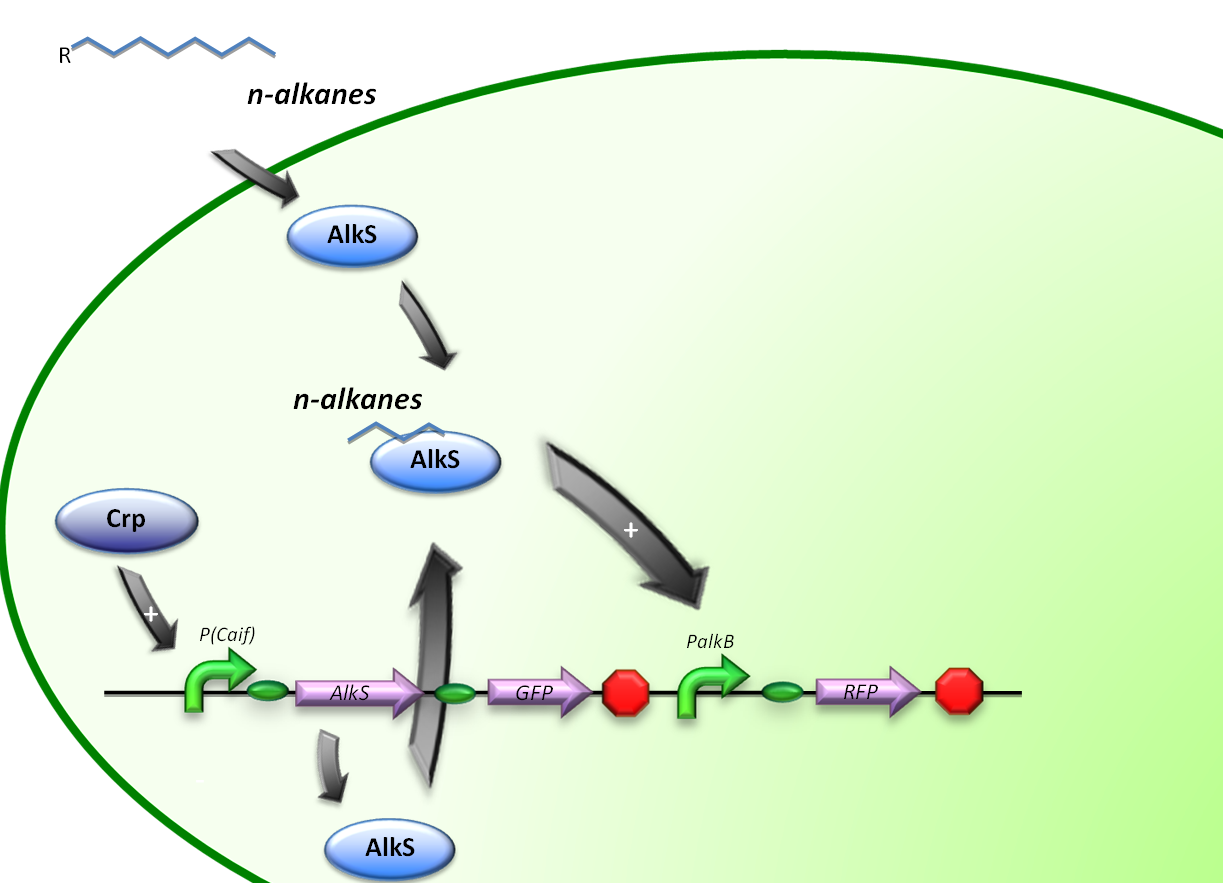
Control by Crp
The expression of alkane degrading genes in our second E.coli regulatory system will be regulated by making the alkane-degrading genes sensitive to Crp. The global regulator protein Crp binds to regions of promoters known to activate genes involved in the degradation of non-glucose carbon sources, in this way activating the genes downstream of it.
For the induction of alkane-degradation genes in this construct the BioBrick consisted out of the P(CaiF) promoter, AlkS gene and PalkB gene. (figure 3)
BBa_K398331: P(Caif)-RBS-GFP-TT
Because of the limited amount of time, we just succeeded on the construction of a measurement device in order to characterize pCaiF. An natural occurring E. coli promotor. We hope that other teams will use these sequence in the future for several reasons, among them the necessity of well characterized promoters in the catalog.
View this part in the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398331 part registry]
References
- Canosa, I., J. M. Sanchez-Romero, et al. A positive feedback mechanism controls expression of AlkS, the transcriptional regulator of the Pseudomonas oleovorans alkane degradation pathway. Molecular Microbiology 35(4): 791-799 (2000)
- Moreno, R., A. Ruiz-Manzano, et al. The Pseudomonas putida Crc global regulator is an RNA binding protein that inhibits translation of the AlkS transcriptional regulator. Molecular Microbiology 64(3): 665-675 (2007)
- van Beilen, J. B., S. Panke, et al. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology-Sgm 147: 1621-1630 (2001)
- Rojo, F. (2009), Degradation of alkanes by bacteria. Environmental Microbiology, 11: 2477–2490. doi: 10.1111/j.1462-2920.2009.01948.x

 "
"