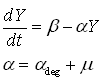Team:TU Delft/Project/rbs-characterization/characterization
From 2010.igem.org
(→Characterization) |
|||
| Line 1: | Line 1: | ||
| - | ==Characterization== | + | ===Characterization=== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | == Plots of measurements == | ||
| + | The RBS strength defines how much of a protein is produced compared to a reference RBS sequence. However, RBS characterization measurements only include current protein level (GFP measurements) and current biomass concentration (Absorption measurements). During measurements, the protein concentration is also influenced by other factors: | ||
| + | * The rapid growth of the bacteria will dilute the protein considerably | ||
| + | * Proteins degrade over time. | ||
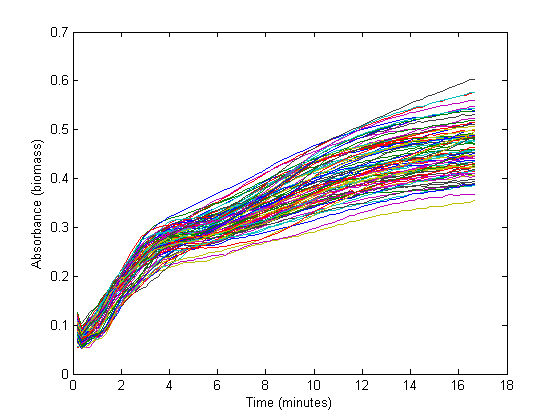
| - | + | From the biomass, or optical density (OD), graph below, it can be seen that the fastest growth occurs from 50 minutes until about 3 hours into the experiment. Within this timespan, it is assumed that growth is exponential, and growth rate can be calculated. The graph below shows 72 growth curves. 12 seperate wells were used for every RBS sequence. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[Image:Tud2010_RBS_OD.png]] | |
| - | + | == Dilution decreasing GFP concentration == | |
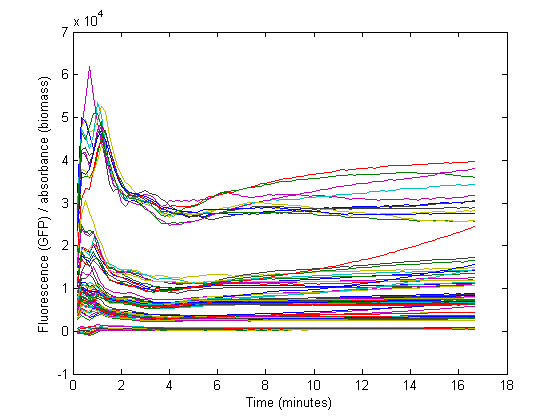
| + | At the start of our measurements, the GFP protein has reached a steady state high concentration. | ||
| + | The specific GFP will decrease during growth because of dilution, and this needs to be accounted for when calculating the RBS strength. | ||
| + | [[Image:TUD2010_Gfpod_all.png]] | ||
| - | == | + | == Protein production model == |
| + | As said, the protein production model needs to take dilution into account. Since the used GFP is a very stable protein, the degradation term is negligible compared to dilution. | ||
| - | + | [[Image:RBS_Expression_model.PNG]] | |
| - | + | In above formula: | |
| + | * Y is the GFP concentration | ||
| + | * Beta is the production rate | ||
| + | * Alpha is the dilution+degradation rate | ||
| - | + | Converting this equation to an explicit form results in: | |
| + | |||
| + | |||
| + | |||
| + | Go to [[Team:TU_Delft/Project/rbs-characterization/results|results]] for measured RBS strengths | ||
| + | |||
| + | == References == | ||
| + | ;Kelly 2009 | ||
| + | :Jason R. Kelly, Adam J. Rubin, Joseph H. Davis, ''et al'' (March 2009). "[http://dx.doi.org/10.1186/1754-1611-3-4 Measuring the activity of BioBrick promoters using an in vivo reference standard]". ''Journal of Biological Engineering'' '''3''': 4 | ||
Revision as of 14:12, 7 October 2010
Contents |
Characterization
Plots of measurements
The RBS strength defines how much of a protein is produced compared to a reference RBS sequence. However, RBS characterization measurements only include current protein level (GFP measurements) and current biomass concentration (Absorption measurements). During measurements, the protein concentration is also influenced by other factors:
- The rapid growth of the bacteria will dilute the protein considerably
- Proteins degrade over time.
From the biomass, or optical density (OD), graph below, it can be seen that the fastest growth occurs from 50 minutes until about 3 hours into the experiment. Within this timespan, it is assumed that growth is exponential, and growth rate can be calculated. The graph below shows 72 growth curves. 12 seperate wells were used for every RBS sequence.
Dilution decreasing GFP concentration
At the start of our measurements, the GFP protein has reached a steady state high concentration. The specific GFP will decrease during growth because of dilution, and this needs to be accounted for when calculating the RBS strength.
Protein production model
As said, the protein production model needs to take dilution into account. Since the used GFP is a very stable protein, the degradation term is negligible compared to dilution.
In above formula:
- Y is the GFP concentration
- Beta is the production rate
- Alpha is the dilution+degradation rate
Converting this equation to an explicit form results in:
Go to results for measured RBS strengths
References
- Kelly 2009
- Jason R. Kelly, Adam J. Rubin, Joseph H. Davis, et al (March 2009). "Measuring the activity of BioBrick promoters using an in vivo reference standard". Journal of Biological Engineering 3: 4
 "
"