Team:TU Delft/27 juli 2010 content
From 2010.igem.org
(Difference between revisions)
| Line 13: | Line 13: | ||
|- | |- | ||
|1 | |1 | ||
| - | |1.5 μg AlkS | + | |1.5 μg AlkS in pSB1C3 |
|XbaI | |XbaI | ||
|SpeI | |SpeI | ||
| Line 21: | Line 21: | ||
|- | |- | ||
|2 | |2 | ||
| - | |1.5 μg AlkS | + | |1.5 μg AlkS in pSB1C3 |
|XbaI | |XbaI | ||
|SpeI | |SpeI | ||
| Line 29: | Line 29: | ||
|- | |- | ||
|3 | |3 | ||
| - | |1.0 μg B0015 | + | |1.0 μg B0015 in pSB1AK3 |
|EcoRI | |EcoRI | ||
|XbaI | |XbaI | ||
| | | | ||
|Buffer 2 (BioLabs) | |Buffer 2 (BioLabs) | ||
| - | |’E X’- pSB1C3 | + | |’E X’- B0015-pSB1C3 |
|- | |- | ||
|4 | |4 | ||
| - | |1.2 μg | + | |1.2 μg E0422 in pS-B1A2 |
|EcoRI | |EcoRI | ||
|XbaI | |XbaI | ||
| | | | ||
|Buffer 2 (BioLabs) | |Buffer 2 (BioLabs) | ||
| - | |‘E X’- pSB1C3 | + | |‘E X’- E0422-pSB1C3 |
|- | |- | ||
|} | |} | ||
| Line 63: | Line 63: | ||
|- | |- | ||
|1 | |1 | ||
| - | |Marker | + | |Marker |
| | | | ||
| | | | ||
Revision as of 13:03, 28 July 2010
Digestion
Some BioBricks are in production. Digestion reactions:
| # | Sample | Enzyme 1 | Enzyme 2 | Enzyme 3 | Buffer | Needed fragment |
| 1 | 1.5 μg AlkS in pSB1C3 | XbaI | SpeI | Buffer 2 + BSA (BioLabs) | ’X - AlkS - S’ | |
| 2 | 1.5 μg AlkS in pSB1C3 | XbaI | SpeI | NCoI | Buffer 2 (BioLabs) | ’X - AlkS - S’ |
| 3 | 1.0 μg B0015 in pSB1AK3 | EcoRI | XbaI | Buffer 2 (BioLabs) | ’E X’- B0015-pSB1C3 | |
| 4 | 1.2 μg E0422 in pS-B1A2 | EcoRI | XbaI | Buffer 2 (BioLabs) | ‘E X’- E0422-pSB1C3 |
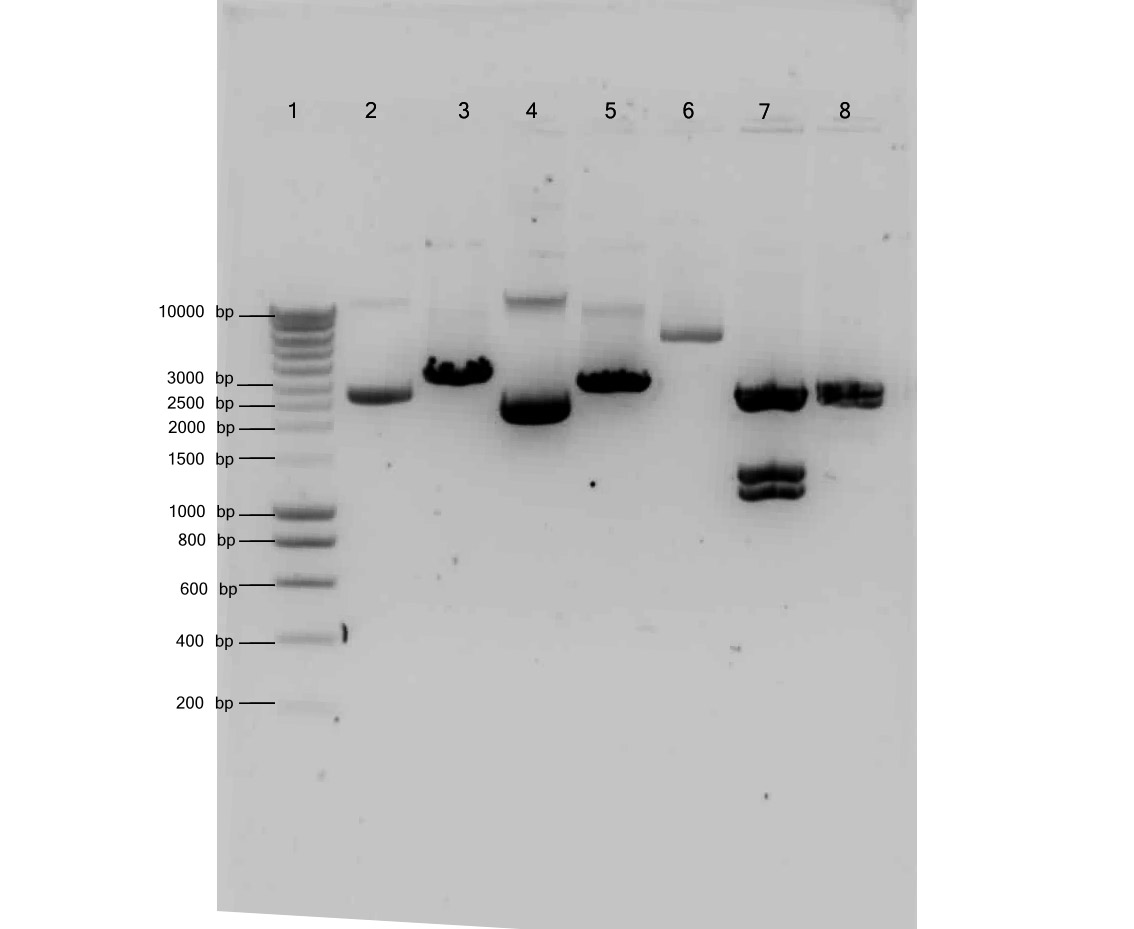
Results of the digestion on 1% agarose gel
Title figure:
1% Agarose of digestion check
gel runned at 100V for 1 hour
loaded 4 μL of marker and 10 μL sample + 2 μL loadingbuffer
Lane description:
| # | Description | Expected Length (bp) | Status |
| 1 | Marker | ||
| 2 | Undigested B0015 in pSB1AK3 | 3318 | ✓ |
| 3 | Digested B0015 in pSB1AK3 + EcoRI + XBaI | 3303, 15 | ✓ |
| 4 | Undigested E0422 in pSB1A2 | 2996 | ✓ |
| 5 | Digested E0422 in pSB1A2 + EcoRI + XBaI | 2981, 15 | ✓ |
| 6 | Undigested AlkS in pSB1C3 | 4999 | ✓ |
| 7 | Digested AlkS in pSB1C3 + EcoRI + SpeI + NCoI | 2673, 1249, 1077 | ✓ |
| 8 | Digested AlkS in pSB1C3 + EcoRI + SpeI | 2673, 2326 | ✓ |
Conclusion: All the plasmids were digested as expected
 "
"