Team:TU Delft/22 July 2010 content
From 2010.igem.org
(Difference between revisions)
(→Alkane degradation) |
|||
| Line 1: | Line 1: | ||
| + | =Lab work= | ||
| + | |||
==Alkane degradation== | ==Alkane degradation== | ||
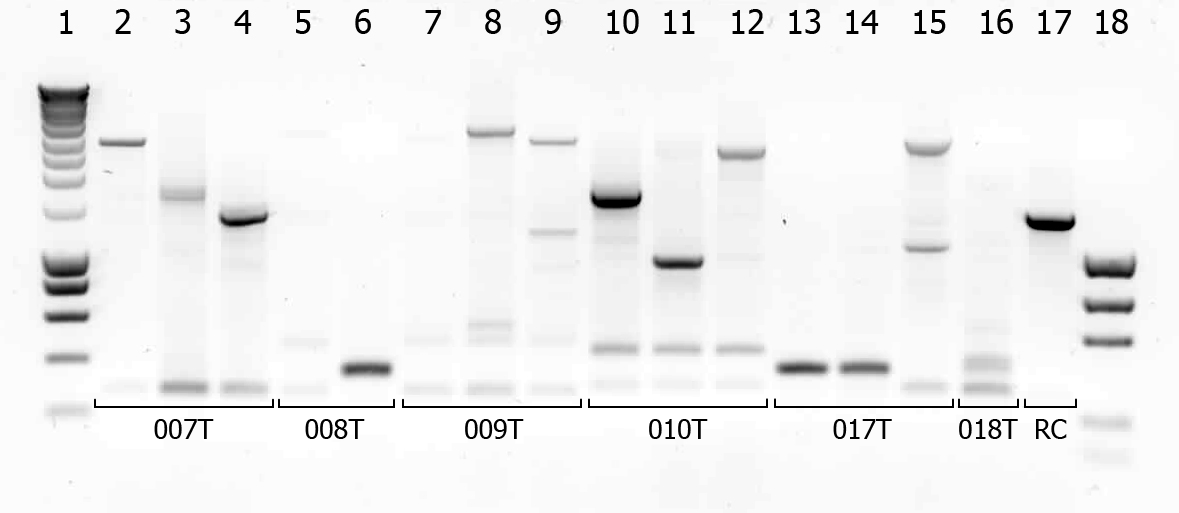
There were some colonies on [https://2010.igem.org/Team:TU_Delft#/blog?blog=19_July_2010 Tuesday's] plates! We had left the plates @ 37°C yesterday after having seen that there were no colonies. When checking this morning on all plates (except the negative control) there were a few colonies! (2-50 colonies). Chances are it's not what we're looking for, but maybe they are good transformants... to check we will do a [[Team:TU_Delft/protocols/colony PCR|colony PCR]]. | There were some colonies on [https://2010.igem.org/Team:TU_Delft#/blog?blog=19_July_2010 Tuesday's] plates! We had left the plates @ 37°C yesterday after having seen that there were no colonies. When checking this morning on all plates (except the negative control) there were a few colonies! (2-50 colonies). Chances are it's not what we're looking for, but maybe they are good transformants... to check we will do a [[Team:TU_Delft/protocols/colony PCR|colony PCR]]. | ||
| - | [[Image:TUDelft_scPCR_22-07.png|550px|thumb|left|1% Agarose gel of | + | [[Image:TUDelft_scPCR_22-07.png|550px|thumb|left|1% Agarose gel of colony PCR. Gel runned at 100V for 1 hour. Of all samples 5 μL + 1 μL loadingbuffer was loaded. 5 μL was loaded of marker.]] |
{| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
|'''#''' | |'''#''' | ||
|'''Description''' | |'''Description''' | ||
|'''Expected length (bp)''' | |'''Expected length (bp)''' | ||
| + | |'''Primers''' | ||
| + | |'''Status''' | ||
| + | |'''Remarks''' | ||
|- | |- | ||
|1 | |1 | ||
| - | |[https://2010.igem.org/Image:TU_Delft_SmartLadder.jpg SmartLadder] | + | |[https://2010.igem.org/Image:TU_Delft_SmartLadder.jpg SmartLadder] |
|n/a | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
|- | |- | ||
| - | |2 | + | |2 |
| - | |007T | + | |Transformant #1 of ligation mix 007T |
|1616 | |1616 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
|- | |- | ||
| - | |5-6 | + | |3 |
| - | |008T | + | |Transformant #2 of ligation mix 007T |
| - | |551 | + | |1616 |
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |4 | ||
| + | |Transformant #3 of ligation mix 007T | ||
| + | |1616 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |5 | ||
| + | |Transformant #1 of ligation mix 008T | ||
| + | |551 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |6 | ||
| + | |Transformant #2 of ligation mix 008T | ||
| + | |551 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |7 | ||
| + | |Transformant #1 of ligation mix 009T | ||
| + | |551 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
|- | |- | ||
| - | | | + | |8 |
| - | |009T | + | |Transformant #1 of ligation mix 009T |
|560 | |560 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
|- | |- | ||
| - | |10-12 | + | |9 |
| - | |010T | + | |Transformant #1 of ligation mix 009T |
| + | |560 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |10 | ||
| + | |Transformant #1 of ligation mix 010T | ||
| + | |1657 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |11 | ||
| + | |Transformant #1 of ligation mix 010T | ||
| + | |1657 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |12 | ||
| + | |Transformant #1 of ligation mix 010T | ||
|1657 | |1657 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
|- | |- | ||
| - | |13 | + | |13 |
| - | |017T | + | |Transformant #1 of ligation mix 017T |
|1130 | |1130 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |14 | ||
| + | |Transformant #1 of ligation mix 017T | ||
| + | |1130 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |15 | ||
| + | |Transformant #1 of ligation mix 017T | ||
| + | |1130 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
|- | |- | ||
|16 | |16 | ||
| - | |018T | + | |Transformant #1 of ligation mix 018T |
|1874 | |1874 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
|- | |- | ||
|17 | |17 | ||
| - | |Red colony | + | |Transformant #1 of Red colony |
| - | | | + | |1360 |
| + | |G00100 + G00101 | ||
| + | | | ||
| + | | | ||
|} | |} | ||
A number of colonies look promising! To check if they really are the BioBricks we want, tomorrow we will do a plasmid isolation with the cultures of lane 2, 4, 6, 7, 9 and 14. We will cut the isolated plasmids with various restriction enzymes and analyze the digestion products on gel. | A number of colonies look promising! To check if they really are the BioBricks we want, tomorrow we will do a plasmid isolation with the cultures of lane 2, 4, 6, 7, 9 and 14. We will cut the isolated plasmids with various restriction enzymes and analyze the digestion products on gel. | ||
Revision as of 19:25, 1 August 2010
Lab work
Alkane degradation
There were some colonies on Tuesday's plates! We had left the plates @ 37°C yesterday after having seen that there were no colonies. When checking this morning on all plates (except the negative control) there were a few colonies! (2-50 colonies). Chances are it's not what we're looking for, but maybe they are good transformants... to check we will do a colony PCR.
| # | Description | Expected length (bp) | Primers | Status | Remarks |
| 1 | SmartLadder | n/a | n/a | n/a | |
| 2 | Transformant #1 of ligation mix 007T | 1616 | G00100 + G00101 | ||
| 3 | Transformant #2 of ligation mix 007T | 1616 | G00100 + G00101 | ||
| 4 | Transformant #3 of ligation mix 007T | 1616 | G00100 + G00101 | ||
| 5 | Transformant #1 of ligation mix 008T | 551 | G00100 + G00101 | ||
| 6 | Transformant #2 of ligation mix 008T | 551 | G00100 + G00101 | ||
| 7 | Transformant #1 of ligation mix 009T | 551 | G00100 + G00101 | ||
| 8 | Transformant #1 of ligation mix 009T | 560 | G00100 + G00101 | ||
| 9 | Transformant #1 of ligation mix 009T | 560 | G00100 + G00101 | ||
| 10 | Transformant #1 of ligation mix 010T | 1657 | G00100 + G00101 | ||
| 11 | Transformant #1 of ligation mix 010T | 1657 | G00100 + G00101 | ||
| 12 | Transformant #1 of ligation mix 010T | 1657 | G00100 + G00101 | ||
| 13 | Transformant #1 of ligation mix 017T | 1130 | G00100 + G00101 | ||
| 14 | Transformant #1 of ligation mix 017T | 1130 | G00100 + G00101 | ||
| 15 | Transformant #1 of ligation mix 017T | 1130 | G00100 + G00101 | ||
| 16 | Transformant #1 of ligation mix 018T | 1874 | G00100 + G00101 | ||
| 17 | Transformant #1 of Red colony | 1360 | G00100 + G00101 |
A number of colonies look promising! To check if they really are the BioBricks we want, tomorrow we will do a plasmid isolation with the cultures of lane 2, 4, 6, 7, 9 and 14. We will cut the isolated plasmids with various restriction enzymes and analyze the digestion products on gel.
 "
"