Team:Stockholm/Modelling
From 2010.igem.org
| Line 1: | Line 1: | ||
| - | |||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
__TOC__ | __TOC__ | ||
| Line 13: | Line 12: | ||
# Cell adhesion | # Cell adhesion | ||
# Antigen presenting | # Antigen presenting | ||
| + | |||
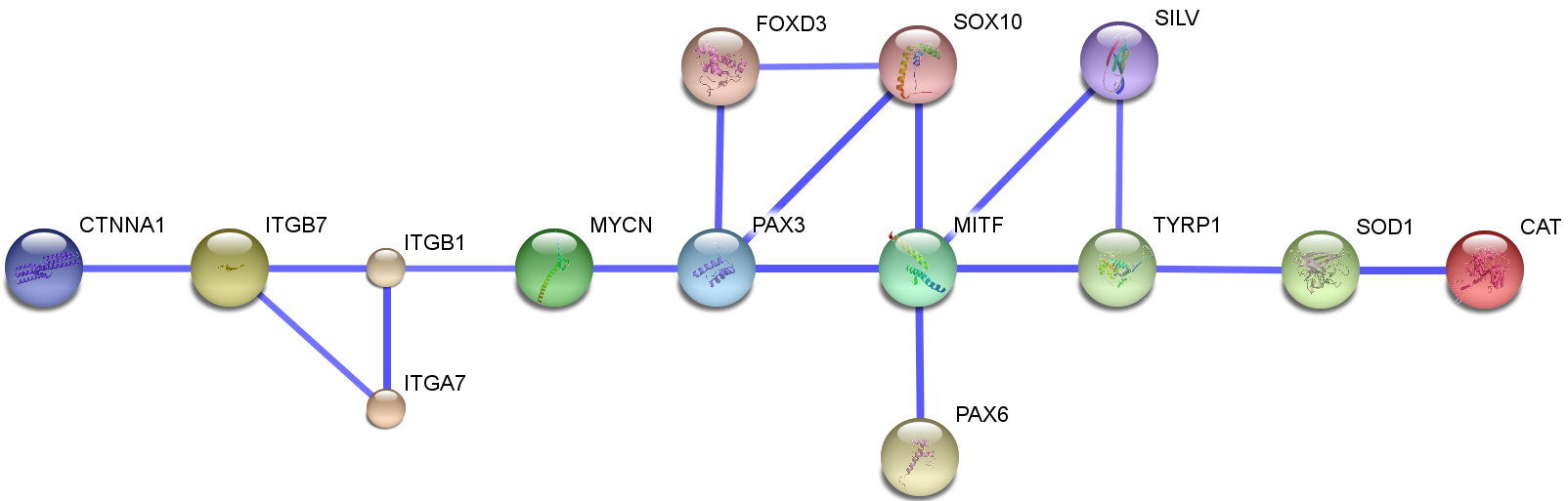
| + | In fig. 1, a very summarized regulatory network of some candidate genes are shown. We intentionally chose interactions that we thought there are enough evidences for it available, to make it more convenient to judge its truth. | ||
---- | ---- | ||
Revision as of 17:47, 1 September 2010
Wet-lab members are working on several proteins. The idea behind why we chose these proteins was somehow vague, because of low amount of information we had on our wiki page. In this section we try to explain the reasons and ideas behind choosing those specific proteins.
As a starting point, we began with the article Strömberg S et al. (2008). In this paper they identified some 859 genes as differentially regulated genes in melanocytes. These genes can be classified in several groups based on their possible role (Strömberg et al. 2008). These groups are:
- Developing melanocytes
- Melanin synthesis and delivering it using melanosomes
- Cell adhesion
- Antigen presenting
In fig. 1, a very summarized regulatory network of some candidate genes are shown. We intentionally chose interactions that we thought there are enough evidences for it available, to make it more convenient to judge its truth.
out of those genes, it seemed that MITF is one of the most regulated and regulating genes in vitiligo disease. This was the first block of the map. . The MITF promoter is targeted by several transcription factors that are important in neural-crest development and signaling. Transcription factors implicated in the regulation of the MITF promoter include PAX3, cAMP-responsive element binding protein (CREB), SOX10, LEF1 (also known as TCF), one cut domain 2 (ONECUT-2) and MITF itself, Fig. 2 (Levy et al., 2006). It also regulates both the survival and differentiation of melanocytes, and enzymes which are necessary for melanin production. (Levy et al., 2006; T.J. Hemesath et al., 1994; N.J. Bentley et al., 1994; K. Yasumoto et al., 1994; C. Bertolotto et al., 1998). So, the regulation of multiple pigmentation and differentiation related genes by MITF (Levy et al., 2006) convinced us that MITF is a central regulator of melanogenesis.
There are evidences for the accumulation of H2O2 in vitiliginous skin (K. U. Schallreuter et al., 1999, 2001, 2006) and low levels of SOD and CAT (A. Jalel et. al., 2008; Koca R et. al., 2004; K. U. Schallreuter et al. 1991; Maresca V et. al. , 1997; ). It was also shown that calcium uptake is defective in vitiliginous skin in keratocytes (K. U. Schallreuter et al. 1988) as well as Melanocytes(K.U. Schallreuter et al., 1996), later the effect of accumulation of H2O2 in the epidermis of patients with vitiligo which leads to disruption calcium homeostasis in the skin was observed (K. U. Schallreuter et al. ,2007). This suggests that an oxidative stress is a pathogenic event in the degeneration of melanocytes (Strömberg S et al. ,2008).
We hypothesized that extra levels of SOD could improve the Melanocytes survivability and help re-pigmentation.
When thinking about re-pigmentation in vitiliginous skin, the first thing comes to mind is lack of melanin. There are evidences that melanocytes are still available in depigmented epidermis of patients with vitiligo even after 25 years (Tobin DJ et. al., 2000). So if melanocytes can't deliver melanin to keratinocytes or they are producing very low amount, it is possible to produce melanin using synthetic biology and deliver it to help re-pigmentation.
 "
"