Team:Stockholm/3 September 2010
From 2010.igem.org
(→Andreas) |
m |
||
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
| + | |||
==Andreas== | ==Andreas== | ||
===Transfer of m-yCCS into pEX & Cloning of N-CPPs=== | ===Transfer of m-yCCS into pEX & Cloning of N-CPPs=== | ||
| Line 47: | Line 48: | ||
Too large bands for all CPP clones to be correct, which is strange. New cloning will be attempted. | Too large bands for all CPP clones to be correct, which is strange. New cloning will be attempted. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
===Cloning of N-CPPs into pSB1C3=== | ===Cloning of N-CPPs into pSB1C3=== | ||
| Line 115: | Line 95: | ||
Difficult to interpret results, since the source plasmid size is unknown. However, it looks like the digested sample presents a shorter fragment compared to the undigested. On the other hand, it also looks like the size difference is too big to have resulted from just excising the ≈300 bp N-CPP cluster.<br /> | Difficult to interpret results, since the source plasmid size is unknown. However, it looks like the digested sample presents a shorter fragment compared to the undigested. On the other hand, it also looks like the size difference is too big to have resulted from just excising the ≈300 bp N-CPP cluster.<br /> | ||
Also strange is that it looks like there are two bands present in the undigested sample. | Also strange is that it looks like there are two bands present in the undigested sample. | ||
| + | |||
| + | ===Cloning of SOD⋅His into pMA=== | ||
| + | ====Sequencing results==== | ||
| + | *SH1: pMA.SOD⋅His 1 ([[media:SH1_premix.txt|fasta]]) | ||
| + | *SH2: pMA.SOD⋅His 2 ([[media:SH2_premix.txt|fasta]]) | ||
| + | |||
| + | Ran nucleotide Blasts (Blastn) to verify the sequencing results ([[media:Blastn_SH1_premix-pMA.SOD*His_3sep.txt|SH1 Blastn]] / [[media:Blastn_SH2_premix-pMA.SOD*His_3sep.txt|SH2 Blastn]]). Both Blasts revealed three identical silent mutations within the His tag. However, since they don't affect the amino acid sequence, and don't result in any rare codons, no action will be needed. | ||
| + | |||
| + | == Mimmi == | ||
| + | |||
| + | === Saftey issues on the home page === | ||
| + | |||
| + | *Making a first layout to answer the saftey questions... | ||
| + | |||
| + | |||
| + | === pMA.his.SOD === | ||
| + | |||
| + | *Plasmid prep. following E.Z.N.A. plasmid mini kit protocol 1 | ||
| + | **Wash two times with DNA wash buffer | ||
| + | **Eluate in 50µl sH<sub>2</sub>O | ||
| + | |||
| + | |||
| + | '''DNA concentration''' | ||
| + | |||
| + | 266ng/µl | ||
| + | |||
| + | |||
| + | |||
| + | === MITF-M === | ||
| + | |||
| + | ==== Colony PCR ==== | ||
| + | |||
| + | Fermentas Pfu | ||
| + | |||
| + | {| | ||
| + | ! Mix | ||
| + | | (µl) | ||
| + | | x5 | ||
| + | | rowspan="9" width="100" | | ||
| + | ! Primers | ||
| + | | rowspan="9" width="100" | | ||
| + | ! colspan="2" | conditions | ||
| + | | rowspan="3" | | ||
| + | |- | ||
| + | | sH<sub>2</sub>O | ||
| + | | 17 | ||
| + | | 85 | ||
| + | | pSB_VF2 | ||
| + | ! time | ||
| + | ! °C | ||
| + | |- | ||
| + | | 10x buffer | ||
| + | | 2.5 | ||
| + | | 12.5 | ||
| + | | pSB_VR | ||
| + | | 2m | ||
| + | | 95 | ||
| + | |- | ||
| + | | dNTP | ||
| + | | 2.5 | ||
| + | | 12.5 | ||
| + | | rowspan="7" | | ||
| + | | 30s | ||
| + | | 95 | ||
| + | | ) | ||
| + | |- | ||
| + | | F primer | ||
| + | | 1 | ||
| + | | 5 | ||
| + | | 30s | ||
| + | | 55 | ||
| + | | > 30 cycles | ||
| + | |- | ||
| + | | R primer | ||
| + | | 1 | ||
| + | | 5 | ||
| + | | 2m40s | ||
| + | | 72 | ||
| + | | ) | ||
| + | |- | ||
| + | | DNA | ||
| + | | 0.5 | ||
| + | | 5x0.5 | ||
| + | | 10m | ||
| + | | 72 | ||
| + | | rowspan="2" | | ||
| + | |- | ||
| + | | Pfu pol | ||
| + | | 0.5 | ||
| + | | 2.5 | ||
| + | | oo | ||
| + | | 10 | ||
| + | |- | ||
| + | | align="right" | tot | ||
| + | | 25µl | ||
| + | | 125µl | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | === pMA.his.SOD === | ||
| + | |||
| + | ==== Digestion ==== | ||
| + | |||
| + | {| | ||
| + | ! Mix | ||
| + | | (µl) | ||
| + | | (µl) | ||
| + | | rowspan="7" width="100" | | ||
| + | ! colspan="2" | Conditions | ||
| + | | rowspan="7" width="100" | | ||
| + | | [pMA.SOD.his] = 246ng/µl -> ~2µg = 8µl | ||
| + | |- | ||
| + | | DNA | ||
| + | | 8 | ||
| + | | 7.5 | ||
| + | ! Time | ||
| + | ! °C | ||
| + | | [pMA.his.SOD] = 280ng/µl -> ~2µg = 7.5µl | ||
| + | |- | ||
| + | | s<sub>2</sub>O | ||
| + | | 17 | ||
| + | | 17.5 | ||
| + | | 30m | ||
| + | | 37 | ||
| + | | [pSB1C3] = 148ng/µl -> ~1.2µg = 8µl | ||
| + | |- | ||
| + | | 10x buffer | ||
| + | | 3 | ||
| + | | 3 | ||
| + | | 20m | ||
| + | | 65 | ||
| + | | rowspan="4" | | ||
| + | |- | ||
| + | | XbaI | ||
| + | | 1 | ||
| + | | 1 | ||
| + | | rowspan="3" colspan="2" | | ||
| + | |- | ||
| + | | PstI | ||
| + | | 1 | ||
| + | | 1 | ||
| + | |- | ||
| + | | align="right" | tot | ||
| + | | 3x30µl | ||
| + | |} | ||
| + | |||
| + | |||
| + | ==== Gel ==== | ||
| + | |||
| + | |||
| + | [[Image:2010-09-03_SOD.his_+_his.SOD_+_pSB1C3_X+P.jpg|100px|thumb|left|]] | ||
| + | {| | ||
| + | ! well | ||
| + | ! sample | ||
| + | |- | ||
| + | | 1 | ||
| + | | 1kb ladder | ||
| + | |- | ||
| + | | 2 | ||
| + | | pMA.SOD.his | ||
| + | |- | ||
| + | | 3 | ||
| + | | pMA.his.SOD | ||
| + | |- | ||
| + | | 4 | ||
| + | | pSB1C3.RFP | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | *Cut gel bands | ||
| + | |||
| + | *Save in fridge | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 10:59, 26 October 2010
Contents |
Andreas
Transfer of m-yCCS into pEX & Cloning of N-CPPs
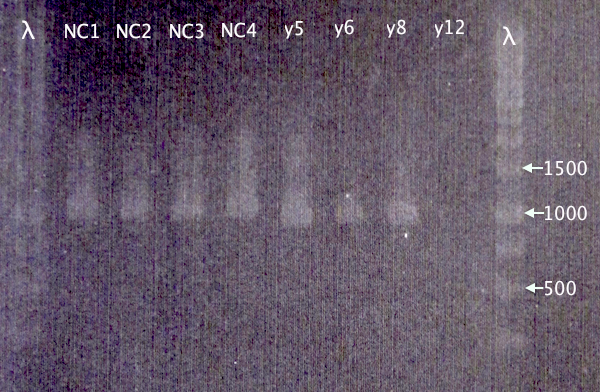
Restreak results from 2/9 revealed four white (positive) clones. These were picked for colony PCR (y5, y6, y8 and y12).
Also picked four colonies from N-CPP plate from 30/8 (NC1, NC2, NC3 and NC4).
Colony PCR
| PCR tubes | |
|---|---|
| dH2O | 16.2 |
| DreamTaq buffer | 2 |
| 10 mM dNTPs | 0.4 |
| pEXf | 0.4 |
| pEXr | 0.4 |
| DNA | 0.5 |
| DreamTaq pol. | 0.08 |
| Total | 20 μl |
Gel verification
1 % agarose, 80 V
Expected bands
- pSB1C3.N-CPPs: 365 bp (TAT), 374 bp (LMWP), 395 bp (Tra10)
- pEX.m-yCCS: 963 bp
Results
Correct bands for pEX.m-yCCS clones 5, 6 and 8. No band for y12. 5 & 8 will later be selected for plasmid prep.
Too large bands for all CPP clones to be correct, which is strange. New cloning will be attempted.
Cloning of N-CPPs into pSB1C3
Re-ligation of 30/8 digestion.
Ligation
[Dig. pSB1C3 X+A EXTR 1] = 13.72 ng/μl [Dig. N-CPP X+A] = 66.6 ng/μl
| Ligation mix | |
|---|---|
| Vector DNA | 6 |
| Insert DNA | 9 |
| 5X Rapid Ligation buf. | 4 |
| dH2O | 0 |
| T4 DNA ligase | 1 |
| Total | 20 μl |
Transformation
Standard transformation procedures.
- 2 μl ligation mix†
- 5 μl ligation mix
†: Possibly contaminated due to non-sterile transformation.
Gel verification of digestion sample
- NCPP: undigested N-CPP cluster plasmid
- Dig NCPP: digested N-CPP cluster plasmid, XbaI and AgeI
1 % agarose, 100 V
Results
Difficult to interpret results, since the source plasmid size is unknown. However, it looks like the digested sample presents a shorter fragment compared to the undigested. On the other hand, it also looks like the size difference is too big to have resulted from just excising the ≈300 bp N-CPP cluster.
Also strange is that it looks like there are two bands present in the undigested sample.
Cloning of SOD⋅His into pMA
Sequencing results
Ran nucleotide Blasts (Blastn) to verify the sequencing results (SH1 Blastn / SH2 Blastn). Both Blasts revealed three identical silent mutations within the His tag. However, since they don't affect the amino acid sequence, and don't result in any rare codons, no action will be needed.
Mimmi
Saftey issues on the home page
- Making a first layout to answer the saftey questions...
pMA.his.SOD
- Plasmid prep. following E.Z.N.A. plasmid mini kit protocol 1
- Wash two times with DNA wash buffer
- Eluate in 50µl sH2O
DNA concentration
266ng/µl
MITF-M
Colony PCR
Fermentas Pfu
| Mix | (µl) | x5 | Primers | conditions | ||||
|---|---|---|---|---|---|---|---|---|
| sH2O | 17 | 85 | pSB_VF2 | time | °C | |||
| 10x buffer | 2.5 | 12.5 | pSB_VR | 2m | 95 | |||
| dNTP | 2.5 | 12.5 | 30s | 95 | ) | |||
| F primer | 1 | 5 | 30s | 55 | > 30 cycles | |||
| R primer | 1 | 5 | 2m40s | 72 | ) | |||
| DNA | 0.5 | 5x0.5 | 10m | 72 | ||||
| Pfu pol | 0.5 | 2.5 | oo | 10 | ||||
| tot | 25µl | 125µl | ||||||
pMA.his.SOD
Digestion
| Mix | (µl) | (µl) | Conditions | [pMA.SOD.his] = 246ng/µl -> ~2µg = 8µl | |||
|---|---|---|---|---|---|---|---|
| DNA | 8 | 7.5 | Time | °C | [pMA.his.SOD] = 280ng/µl -> ~2µg = 7.5µl | ||
| s2O | 17 | 17.5 | 30m | 37 | [pSB1C3] = 148ng/µl -> ~1.2µg = 8µl | ||
| 10x buffer | 3 | 3 | 20m | 65 | |||
| XbaI | 1 | 1 | |||||
| PstI | 1 | 1 | |||||
| tot | 3x30µl | ||||||
Gel
| well | sample |
|---|---|
| 1 | 1kb ladder |
| 2 | pMA.SOD.his |
| 3 | pMA.his.SOD |
| 4 | pSB1C3.RFP |
- Cut gel bands
- Save in fridge
 |

|
 |

|
 |

|

|

|
 "
"