Team:Stockholm/28 July 2010
From 2010.igem.org
NinaSchiller (Talk | contribs) (→Glycerol stock on CPP and IgG protease) |
NinaSchiller (Talk | contribs) (→Streaking out tyrosinase) |
||
| Line 18: | Line 18: | ||
---- | ---- | ||
| - | ==Streaking out tyrosinase== | + | ===Streaking out tyrosinase=== |
I stroke out tyrosinase which we obatined from iGEM hq in a glycerol stock on a dish containing ampicillin resistance. It was incubated in 37 °C ON. This will be followed tomorrow with a colony-PCR. | I stroke out tyrosinase which we obatined from iGEM hq in a glycerol stock on a dish containing ampicillin resistance. It was incubated in 37 °C ON. This will be followed tomorrow with a colony-PCR. | ||
---- | ---- | ||
| + | |||
==PCR on tyrosinase== | ==PCR on tyrosinase== | ||
Revision as of 18:49, 25 August 2010
Contents |
Nina
Glycerol stock on CPP and IgG protease
I made glycerol stocks on the CPP transportan 10 C version from dish +8ng/ul. Colony number 9 and 40 were added each to a falcon tube containing 12 ml LB and 24 ul ampicillin resistance (50 mg/ml). The tubes were incubated in 37 °C until OD reaches around 0.6. After around 4 hours the OD was about 0.6.
I also made a glycerol stock on IgG protease inserted in the linjerized shipping vector (with chloramphenicol resistance). Colony number 2 was added to a falcon tube containing 12 ml LB and 24 ul chloramphenicol resistance (50 mg/ml). The tubes were incubated in 37 °C until OD reaches around 0.6, which was obtained after around 5 hours.
Glycerol stock:
- 400 ul Glycerol
- 800 ul Bacterial sample
Streaking out tyrosinase
I stroke out tyrosinase which we obatined from iGEM hq in a glycerol stock on a dish containing ampicillin resistance. It was incubated in 37 °C ON. This will be followed tomorrow with a colony-PCR.
PCR on tyrosinase
I ran a PCR on tyrosinase (1ul) from a glycerol stock with the gene's forward and reverse primers. The annealing time was 2 min.
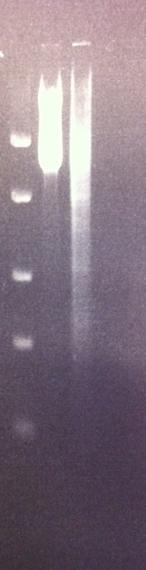
Agarose gel on tyrosinase and IgG protease
Both the PCR product of tyrosinase from todays work and the mini prep of IgG protease in the shipping vector from yesterday's work were run on an agarose gel (2%).
Arrangement on gel:
DNA Ladder: FastRuler™ Middle Range, ready-to-use, 100-5000 bp Fermentas
Unfortunately neither one of the samples did turn out well in the gel. In the case of the IgG protease I believe it shows both an upper band representing coild vectors and a lower band showing the uncoild vector. Therefore I will instead cut the vector with two restriction enzymes called EcoRI and PstI, which I inserted the gene into the vector with. This should present two bands, both the gene IgG protease and the cut vector. This is a good proof that I have my gene in the vector before I send this part in to iGEM hq.
In the case of the tyrosinase I see a lot of unspecific binding, however the sample of tyrosinase I used in the PCR was from a glycerol stock. This can have had a bad effect on the PCR procedure, resulting in the poor result in the tyrosinase lane. Next I will run a tyrosinase colony PCR (withouth any glycerol interfering) with both 2,5 and 3 min annealing time since the gene is long.
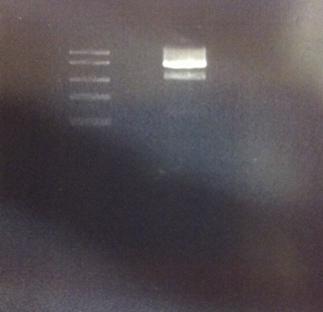
Agarose gel on digested IgG protease in shipping vector
I made a digestion of the IgG protease in the shipping vector with chloramphenicol resistance.
Digestion:
- 20 ul vector
- 1 ul of each restriction enzyme (EcoRI and PstI FastDigest® restriction enzymes fermentas)
- 3 ul buffer (10X FastDigest® Buffer)
- 5 ul H2O
Total volume 30 ul
Incubate in 37 °C for 15 min. Run on agarose gel 1%.
DNA Ladder: FastRuler™ Middle Range, ready-to-use, 100-5000 bp Fermentas.
The result of this gel is good since it shows both the IgG protease gene and the cut vector separated.
CPP ligation on version transportan 10 N
I made a ligation on the CPP transportan 10 version N. Fixa detaljer!
Transformation of IgG protease and CPP
I transformed with minipreped IgG in pEX colony number 18 (tube 1/7-2010) in BL21 for an IPTG treatment tomorrow. As a positive control I also transformed bFGF inserted in its original vector. This had been diluted 30 times, therefore I transformed the bacteria with 1 ul, whereas I transformed with 3 ul of the IgG protease.
I also transformed the CPP Transportan 10 C version into BL21 cells. As a negative control I transformed with an insert (named +8ng/ul) and without an insert (named -8ng/ul).
The transformation method is according to the procedure decribed in protocols.
Note that during the ligation step (don't know the time) of the CPP Johan placed by accident all the tubes in the freezer (-20°C). Therefore after 3 hours we took out the samples into room temp and added additional 0.5 ul ligase T4 and incubated in room temp for 1 h.
Mimmi
over-expression in pEX
- - remaking the comassie-gel
- Heat the samples to 95°C, 3min
- Load 3µl sample to a mini-gel with a 8/1 comb
- Run program 6, comassie gel (15%)
- Run program 2, staining
Site-Directed Mutagenesis
- Deactivate Dpn1
- Heat samples to 80°C, 20min
- Deactivate Dpn1
- Transform into Top10 cells
- Add 1µl to 100µl cells
- Hold on ice 30min
- Heat shock 42°C, 55sec
- Cool down
- Add 900µl LB
- Incubate in 37°C, 1h, ~250rpm
- Johan plated and put it in 37°C ON
- Transform into Top10 cells
 "
"