Team:Stockholm/24 July 2010
From 2010.igem.org
Revision as of 00:24, 1 August 2010 by NinaSchiller (Talk | contribs)
Contents |
Johan
Colony PCR
- A colony PCR on the TAT CCP (N-version) was performed as the result from the last gel electrophoresis (insert date) was just a smear on almost all bands. The reason was most probably too much genomic DNA (too much colony picked), so less of the colonies was picked and the final volume was increased from 25 µl to 50 µl.
- A colony PCR on the LMWP CCP (N- & C-version) was performed as the result from Mimmis gel electrophoresis was just empty bands (insert date).
- TAT: colonies #1-41 +insert, #42-43 -insert.
- LMWP: colonies #1 -insert, #3-24 +insert.
- PCR reaction mix
- 1 µl Pfu polymerase
- 1 µl 10 mM dNTPs
- 3 µl 10 µm f.primer (VF2)
- 3 µl 10 µm r.primer (VR)
- 5 µl Pfu buffer 10x
- 32 µl H2O (miscalculation, should have been 37 µl)
- PCR program
- 98 °C - 2 min
- 35 cycles of
- 98 °C - 10 sec
- 55 °C - 15 sec
- 72 °C - 30 sec
- 72 °C - 5 min
- 4°C - ∞
Nina
Transformation of IgG protease in shipping vector
I transformed 100 ul of competent top 10 cells with 3 ul IgG protease in shipping vector. This was plated on dishes with chloramphenicol resistance.
The transformation method was carried out according to the procedure described in protocols.
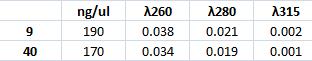
Mini prep of CPP transportan 10 version C
I carried out a mini prep of CPP transportan 10 version C colony numbers 9 and 40.
The mini prep method was carried out according to the procedure described in protocols.
Measuring concentration with spectrophotometer:
 "
"