Team:Stockholm/20 August 2010
From 2010.igem.org
Contents |
Hassan
midnight update, after a fast recovery from sickness!
Version 0.5.1
Version 0.6.1
Mimmi
pMA
Primer dilution
| pMA_VF | 91µl sH2O --> 100µM | 10µl + 90µl sH2O --> 10µM | ||
| pMA_VR | 91µl sH2O --> 100µM | 10µl + 90µl sH2O --> 10µM |
pMA/pEX
verification PCR
| pEX | pMA | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mix | (µl) | X6 | X3 | primers | conditions | ||||
| sH2O | 22.5 | 135 | 67.5 | pEX_VF | time | °C | |||
| F primer | 1 | 6 | 3 | pEX_VR | 2m | 94 | |||
| R primer | 1 | 6 | 3 | pMA_VF | 30s | 94 | ) | ||
| DNA | 0.5 | 5X0.5 | 2X0.5 | pMA_VR | 30s | 60 | > 30 cycles | ||
| tot | 25µl | 250µl | 75µl | 2m30s | 72 | ) | |||
| 10m | 72 | ||||||||
| oo | 10 | ||||||||
Andreas
Transformation results
From 19/8 transformations
- pEX.RFP: Good yield of red (positive) and white (negative) colonies.
- pSB1K3.SOD⋅His: Very few colonies
- pSB1K3.yCCS.His: Very few colonies
- Km probably not feasible with quick transformation.
Colony PCR of pEX.RFP, pEX and pMA.His
Picked four pEX.RFP colonies for PCR verification. Also ran PCR on the pEX and pMA.His clones from 19/8. PCR and gel run by Mimmi (see above).
Results
- pMA.His clone verified for correct insert.
- pEX and pEX.RFP need to be re-run, as they did not result in any bands.
ON cultures
- pMA.His
- pEX (not yet verified)
- pEX.RFP 3 (not yet verified)
3 ml LB + 100 Amp. 30 °C ON.
Plasmid prep
From 19/8 ON cultures
- E.Z.N.A. Plasmid Mini Prep kit.
- 70 μl elution volume
| DNA concentrations | ||
|---|---|---|
| Sample | Conc. [ng/μl] | A260/A280 |
| pSB1K3.BBa_J04450 | 103.8 | 1.94 |
| pEX | 55.52 | 1.89 |
| pMA.His | 85.67 | 1.87 |
Assembly of His⋅SOD/yCCS into pSB1K3
Step I of C-CPP cloning strategy (19/8)
Digestions
[pSB1C3.m-yCCS 1] = 101.2 ng/μl
[pSB1C3.m-SOD 2] = 105.5 ng/μl
| [μl] | pMA.His | pSB1C3.m-yCCS 1 | pSB1C3.m-SOD 1 | Inc.: 37 °C, 0:30 (FD) or 1:30 (NgoMIV) |
|---|---|---|---|---|
| 10X FD buffer | 3 | 3 | 3 | |
| dH2O | 6.7† | 6 | 7 | |
| 2 μg DNA | 23.3 | 20 | 19 | |
| FD EcoRI | 0.5 | – | – | |
| FD PstI | – | 1 | 1 | |
| FD AgeI | 0.5 | – | – | |
| NgoMIV | – | 1 | 1 | |
| 30† | 30 | 30 |
† Miscalculation
Ligations
Without prior enzyme inactivation or DNA purification.
| [μl] | Lig pSB1K3. His⋅SOD | Lig pSB1K3. His⋅yCCS | Vector/insert ratio: 1:3. Inc.: 22 °C, 0:10 |
|---|---|---|---|
| 100 ng vector | 2.2 | 2.2 | |
| His insert | 4.0 | 4.0 | |
| SOD insert | 5.1 | – | |
| yCCS | – | 5.7 | |
| 5X Rapid Lig. buf. | 4 | 4 | |
| dH2O | 2.7 | 2.1 | |
| T4 DNA ligase | 1 | 1 | |
| 20 | 20 |
Enzyme inactivation (not PstI): 80 °C, 20 min.
Transformation
Standard transformation protocol.
- 1 μl ligation mix
- Lig pSB1K3.His⋅SOD
- Lig pSB1K3.His⋅yCCS
- Cells plated onto 50 Km LB agar plates
Nina
Colony PCR on fusion protein
I performed a colony PCR on the dish with fusion proteins.
PCR mix for 5 tubes:
- MgCl 5 ul
- Buffer 5X 50 ul
- dNTP 5 ul
- primer revers for IgG protease 15 ul
- primer VF2 15 ul
- polymerase PjuX7 5 ul
- H2O 150 ul
PCR prgm:
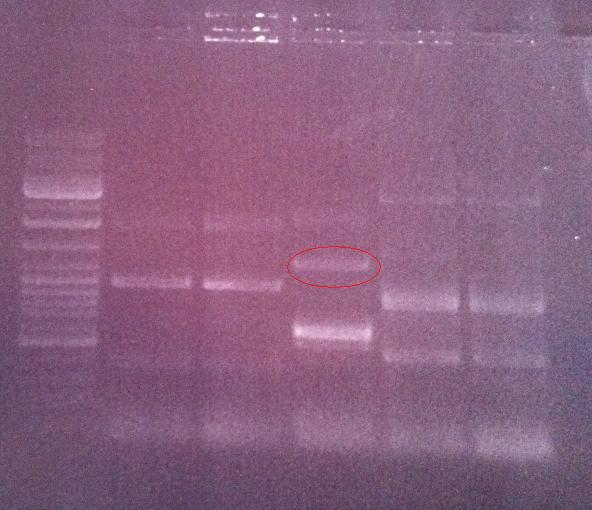
Agarose gel on fusion protein
I ran the PCR product on an 1 % agarose gel 100 V in order to verify that I have bands corresponding to a correct size of the fusion protein.
Ladder: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas
Arrangement on gel:
Lane nr 3 looks like it could represent the wanted band of the fusion protein by protein A and IgG protease, which could be about 1300 nt.
Overnight culture of fusion protein
I inoculated colony nr 3 of the dish with the fusion protein into 12 ml LB and 24 ul chloramphenicol (50 mg/ml). This was incubated in 37 °C in shake overnight.
 "
"