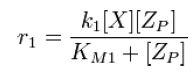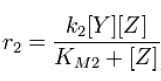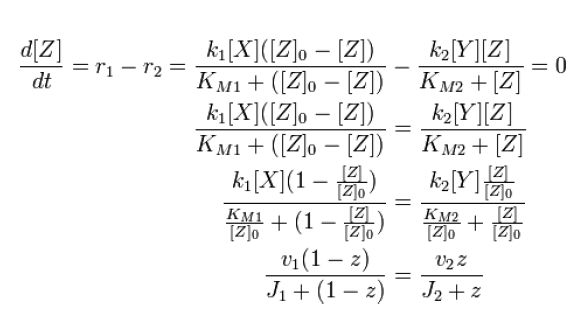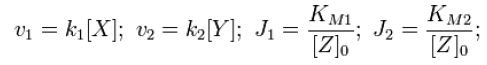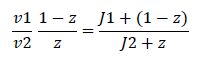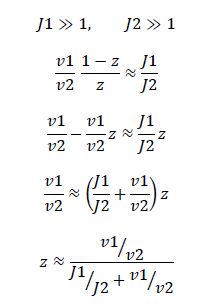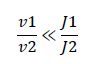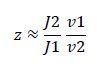|
|
| Line 9: |
Line 9: |
| | | | |
| | | | |
| - | ===sRNA System===
| |
| - |
| |
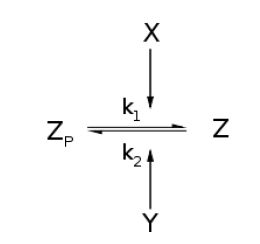
| - | Boolean model for the sRNA system. This is mean to provide a significantly simplified mathematical understanding of the dynamics involved.
| |
| - |
| |
| - | <html>
| |
| - | <head>
| |
| - | <meta http-equiv="Content-Type" content="text/html; charset=utf-8">
| |
| - | <!--
| |
| - | This HTML is auto-generated from an M-file.
| |
| - | To make changes, update the M-file and republish this document.
| |
| - | --><title>iGEM_boolean_sRNA_model</title><meta name="generator" content="MATLAB 7.10"><meta name="date" content="2010-10-27"><meta name="m-file" content="iGEM_boolean_sRNA_model"><style type="text/css">
| |
| - |
| |
| - | body {
| |
| - | background-color: white;
| |
| - | margin:10px;
| |
| - | }
| |
| - |
| |
| - | h1 {
| |
| - | color: #990000;
| |
| - | font-size: x-large;
| |
| - | }
| |
| - |
| |
| - | h2 {
| |
| - | color: #990000;
| |
| - | font-size: medium;
| |
| - | }
| |
| - |
| |
| - | /* Make the text shrink to fit narrow windows, but not stretch too far in
| |
| - | wide windows. */
| |
| - | p,h1,h2,div.content div {
| |
| - | max-width: 600px;
| |
| - | /* Hack for IE6 */
| |
| - | width: auto !important; width: 600px;
| |
| - | }
| |
| - |
| |
| - | pre.codeinput {
| |
| - | background: #EEEEEE;
| |
| - | padding: 10px;
| |
| - | }
| |
| - | @media print {
| |
| - | pre.codeinput {word-wrap:break-word; width:100%;}
| |
| - | }
| |
| - |
| |
| - | span.keyword {color: #0000FF}
| |
| - | span.comment {color: #228B22}
| |
| - | span.string {color: #A020F0}
| |
| - | span.untermstring {color: #B20000}
| |
| - | span.syscmd {color: #B28C00}
| |
| - |
| |
| - | pre.codeoutput {
| |
| - | color: #666666;
| |
| - | padding: 10px;
| |
| - | }
| |
| - |
| |
| - | pre.error {
| |
| - | color: red;
| |
| - | }
| |
| - |
| |
| - | p.footer {
| |
| - | text-align: right;
| |
| - | font-size: xx-small;
| |
| - | font-weight: lighter;
| |
| - | font-style: italic;
| |
| - | color: gray;
| |
| - | }
| |
| - |
| |
| - | </style></head><body><div class="content"><pre class="codeinput">clear <span class="string">all</span>
| |
| - | clc
| |
| - |
| |
| - | <span class="comment">%Constants and Values</span>
| |
| - | trackingNum = 6; <span class="comment">%mRNA_pA, mRNA_pB, sRNA_A, sRNA_B, protein_A, protein_B;</span>
| |
| - | inputNum = 2; <span class="comment">%A, B;</span>
| |
| - | processNum = 6; <span class="comment">%Trx_pA, Trx_sRNA_A, Trx_pB, Trx_sRNA_B, Trl_A, Trl_B;</span>
| |
| - |
| |
| - | <span class="comment">%Cell/Matrix Dimensions</span>
| |
| - | m = 2^trackingNum;
| |
| - | n = 2^inputNum;
| |
| - | c = cell(m+1, n+1);
| |
| - |
| |
| - | trackingMatrix = de2bi(0:m-1);
| |
| - | inputMatrix = de2bi(0:n-1);
| |
| - | processMatrix = zeros(1,processNum);
| |
| - |
| |
| - | <span class="comment">%Populating the Cell (Tracking x Input)</span>
| |
| - | c{1,1} = [0];
| |
| - | <span class="keyword">for</span> j = 1
| |
| - | <span class="keyword">for</span> i = 2:m+1
| |
| - | c{i,j} = trackingMatrix(i-1,:);
| |
| - | <span class="keyword">end</span>
| |
| - | <span class="keyword">end</span>
| |
| - | <span class="keyword">for</span> i = 1
| |
| - | <span class="keyword">for</span> j = 2:n+1
| |
| - | c{i,j} = inputMatrix(j-1,:);
| |
| - | <span class="keyword">end</span>
| |
| - | <span class="keyword">end</span>
| |
| - | <span class="keyword">for</span> i = 2:m+1
| |
| - | <span class="keyword">for</span> j = 2:n+1
| |
| - | c{i,j} = processMatrix;
| |
| - | <span class="keyword">end</span>
| |
| - | <span class="keyword">end</span>
| |
| - |
| |
| - | <span class="comment">%Rules, Acessing, and Changing</span>
| |
| - | <span class="keyword">for</span> j = 2:n+1
| |
| - | A = c{1,j}(1);
| |
| - | B = c{1,j}(2);
| |
| - |
| |
| - | <span class="keyword">for</span> i = 2:m+1
| |
| - | mRNA_pA = c{i,1}(1);
| |
| - | mRNA_pB = c{i,1}(2);
| |
| - | sRNA_A = c{i,1}(3);
| |
| - | sRNA_B = c{i,1}(4);
| |
| - | protein_A = c{i,1}(5);
| |
| - | protein_B = c{i,1}(6);
| |
| - |
| |
| - | <span class="comment">%Trx_pA</span>
| |
| - | <span class="keyword">if</span> A == 1
| |
| - | c{i,j}(1) = 1;
| |
| - | <span class="keyword">end</span>
| |
| - |
| |
| - | <span class="comment">%Trx_pB</span>
| |
| - | <span class="keyword">if</span> B == 1
| |
| - | c{i,j}(2) = 1;
| |
| - | <span class="keyword">end</span>
| |
| - |
| |
| - | <span class="comment">%Trx_sRNA_A</span>
| |
| - | <span class="keyword">if</span> A == 1
| |
| - | c{i,j}(3) = 1;
| |
| - | <span class="keyword">end</span>
| |
| - |
| |
| - | <span class="comment">%Trx_sRNA_B</span>
| |
| - | <span class="keyword">if</span> B == 1
| |
| - | c{i,j}(4) = 1;
| |
| - | <span class="keyword">end</span>
| |
| - |
| |
| - | <span class="comment">%Trl_A</span>
| |
| - | <span class="keyword">if</span> mRNA_pA == 1 && mRNA_pB == 0
| |
| - | c{i,j}(5) = 1;
| |
| - | <span class="keyword">end</span>
| |
| - |
| |
| - | <span class="comment">%Trl_B</span>
| |
| - | <span class="keyword">if</span> mRNA_pB == 1 && mRNA_pA == 0
| |
| - | c{i,j}(6) = 1;
| |
| - | <span class="keyword">end</span>
| |
| - |
| |
| - | <span class="keyword">end</span>
| |
| - | <span class="keyword">end</span>
| |
| - |
| |
| - | <span class="comment">%Display Results (Column by Column)</span>
| |
| - | [nrows,ncols]= size(c);
| |
| - | <span class="comment">%Condense Cell and Display</span>
| |
| - | <span class="keyword">for</span> i = 1:nrows
| |
| - | <span class="keyword">for</span> j = 1:ncols
| |
| - | string = num2str(c{i,j});
| |
| - | l = length(string);
| |
| - | r = 1;
| |
| - | s = 1;
| |
| - | t = 0;
| |
| - |
| |
| - | <span class="keyword">while</span> t ~= 1
| |
| - | <span class="keyword">if</span> r == l
| |
| - | t = 1;
| |
| - | <span class="keyword">end</span>
| |
| - |
| |
| - | noSpacesString(s) = string(r);
| |
| - | r = r+3;
| |
| - | s = s+1;
| |
| - | <span class="keyword">end</span>
| |
| - | c{i,j} = noSpacesString;
| |
| - | <span class="keyword">end</span>
| |
| - | <span class="keyword">end</span>
| |
| - | c(:,:)
| |
| - |
| |
| - | <span class="comment">%Find Steady States and Corresponding Inputs</span>
| |
| - | counter = 1;
| |
| - | <span class="keyword">for</span> i = 2:m+1
| |
| - | <span class="keyword">for</span> j = 2:n+1
| |
| - | <span class="keyword">if</span> c{i,j} == c{i,1}
| |
| - | completeMatrix{counter,1} = c{i,j}; <span class="comment">%Steady state values</span>
| |
| - | completeMatrix{counter,2} = num2str([i-1,j-1]); <span class="comment">%Location (n x m) within results area</span>
| |
| - | completeMatrix{counter,3} = c{1,j}; <span class="comment">%Corresponding input values</span>
| |
| - | counter = counter+1;
| |
| - | <span class="keyword">end</span>
| |
| - | <span class="keyword">end</span>
| |
| - | <span class="keyword">end</span>
| |
| - | SS_mXn_Input = completeMatrix
| |
| - |
| |
| - | time = 0:processNum;
| |
| - | <span class="comment">%plot(time,,time,,time,,time,,time,,time,)</span>
| |
| - |
| |
| - | <span class="comment">%New Cell for Specific Cases</span>
| |
| - | <span class="comment">% d(:,1) = c(:,1);</span>
| |
| - | <span class="comment">% for j = 2:n+1</span>
| |
| - | <span class="comment">% C1 = '0000';</span>
| |
| - | <span class="comment">% C2 = '0011';</span>
| |
| - | <span class="comment">% C3 = '0100';</span>
| |
| - | <span class="comment">% C4 = '0111';</span>
| |
| - | <span class="comment">% C5 = '1000';</span>
| |
| - | <span class="comment">% C6 = '1011';</span>
| |
| - | <span class="comment">% C7 = '1100';</span>
| |
| - | <span class="comment">%</span>
| |
| - | <span class="comment">% a = {C1, C2, C3, C4, C5, C6, C7};</span>
| |
| - | <span class="comment">% for b = 1:1:length(a)</span>
| |
| - | <span class="comment">% if strcmp(c(1,j),a(b)) == 1</span>
| |
| - | <span class="comment">% d(:,b+1) = c(:,j);</span>
| |
| - | <span class="comment">% end</span>
| |
| - | <span class="comment">% end</span>
| |
| - | <span class="comment">% end</span>
| |
| - | <span class="comment">% d</span>
| |
| - | </pre><pre class="codeoutput">
| |
| - | ans =
| |
| - |
| |
| - | '0' '00' '01' '10' '11'
| |
| - | '000000' '000000' '010100' '101000' '111100'
| |
| - | '000001' '000000' '010100' '101000' '111100'
| |
| - | '000010' '000000' '010100' '101000' '111100'
| |
| - | '000011' '000000' '010100' '101000' '111100'
| |
| - | '000100' '000000' '010100' '101000' '111100'
| |
| - | '000101' '000000' '010100' '101000' '111100'
| |
| - | '000110' '000000' '010100' '101000' '111100'
| |
| - | '000111' '000000' '010100' '101000' '111100'
| |
| - | '001000' '000000' '010100' '101000' '111100'
| |
| - | '001001' '000000' '010100' '101000' '111100'
| |
| - | '001010' '000000' '010100' '101000' '111100'
| |
| - | '001011' '000000' '010100' '101000' '111100'
| |
| - | '001100' '000000' '010100' '101000' '111100'
| |
| - | '001101' '000000' '010100' '101000' '111100'
| |
| - | '001110' '000000' '010100' '101000' '111100'
| |
| - | '001111' '000000' '010100' '101000' '111100'
| |
| - | '010000' '000001' '010101' '101001' '111101'
| |
| - | '010001' '000001' '010101' '101001' '111101'
| |
| - | '010010' '000001' '010101' '101001' '111101'
| |
| - | '010011' '000001' '010101' '101001' '111101'
| |
| - | '010100' '000001' '010101' '101001' '111101'
| |
| - | '010101' '000001' '010101' '101001' '111101'
| |
| - | '010110' '000001' '010101' '101001' '111101'
| |
| - | '010111' '000001' '010101' '101001' '111101'
| |
| - | '011000' '000001' '010101' '101001' '111101'
| |
| - | '011001' '000001' '010101' '101001' '111101'
| |
| - | '011010' '000001' '010101' '101001' '111101'
| |
| - | '011011' '000001' '010101' '101001' '111101'
| |
| - | '011100' '000001' '010101' '101001' '111101'
| |
| - | '011101' '000001' '010101' '101001' '111101'
| |
| - | '011110' '000001' '010101' '101001' '111101'
| |
| - | '011111' '000001' '010101' '101001' '111101'
| |
| - | '100000' '000010' '010110' '101010' '111110'
| |
| - | '100001' '000010' '010110' '101010' '111110'
| |
| - | '100010' '000010' '010110' '101010' '111110'
| |
| - | '100011' '000010' '010110' '101010' '111110'
| |
| - | '100100' '000010' '010110' '101010' '111110'
| |
| - | '100101' '000010' '010110' '101010' '111110'
| |
| - | '100110' '000010' '010110' '101010' '111110'
| |
| - | '100111' '000010' '010110' '101010' '111110'
| |
| - | '101000' '000010' '010110' '101010' '111110'
| |
| - | '101001' '000010' '010110' '101010' '111110'
| |
| - | '101010' '000010' '010110' '101010' '111110'
| |
| - | '101011' '000010' '010110' '101010' '111110'
| |
| - | '101100' '000010' '010110' '101010' '111110'
| |
| - | '101101' '000010' '010110' '101010' '111110'
| |
| - | '101110' '000010' '010110' '101010' '111110'
| |
| - | '101111' '000010' '010110' '101010' '111110'
| |
| - | '110000' '000000' '010100' '101000' '111100'
| |
| - | '110001' '000000' '010100' '101000' '111100'
| |
| - | '110010' '000000' '010100' '101000' '111100'
| |
| - | '110011' '000000' '010100' '101000' '111100'
| |
| - | '110100' '000000' '010100' '101000' '111100'
| |
| - | '110101' '000000' '010100' '101000' '111100'
| |
| - | '110110' '000000' '010100' '101000' '111100'
| |
| - | '110111' '000000' '010100' '101000' '111100'
| |
| - | '111000' '000000' '010100' '101000' '111100'
| |
| - | '111001' '000000' '010100' '101000' '111100'
| |
| - | '111010' '000000' '010100' '101000' '111100'
| |
| - | '111011' '000000' '010100' '101000' '111100'
| |
| - | '111100' '000000' '010100' '101000' '111100'
| |
| - | '111101' '000000' '010100' '101000' '111100'
| |
| - | '111110' '000000' '010100' '101000' '111100'
| |
| - | '111111' '000000' '010100' '101000' '111100'
| |
| - |
| |
| - |
| |
| - | SS_mXn_Input =
| |
| - |
| |
| - | '000000' '1 1' '00'
| |
| - | '010101' '22 2' '01'
| |
| - | '101010' '43 3' '10'
| |
| - | '111100' '61 4' '11'
| |
| - |
| |
| - | </pre><p class="footer"><br>
| |
| - | Published with MATLAB® 7.10<br></p></div><!--
| |
| - | ##### SOURCE BEGIN #####
| |
| - | clear all
| |
| - | clc
| |
| - |
| |
| - | %Constants and Values
| |
| - | trackingNum = 6; %mRNA_pA, mRNA_pB, sRNA_A, sRNA_B, protein_A, protein_B;
| |
| - | inputNum = 2; %A, B;
| |
| - | processNum = 6; %Trx_pA, Trx_sRNA_A, Trx_pB, Trx_sRNA_B, Trl_A, Trl_B;
| |
| - |
| |
| - | %Cell/Matrix Dimensions
| |
| - | m = 2^trackingNum;
| |
| - | n = 2^inputNum;
| |
| - | c = cell(m+1, n+1);
| |
| - |
| |
| - | trackingMatrix = de2bi(0:m-1);
| |
| - | inputMatrix = de2bi(0:n-1);
| |
| - | processMatrix = zeros(1,processNum);
| |
| - |
| |
| - | %Populating the Cell (Tracking x Input)
| |
| - | c{1,1} = [0];
| |
| - | for j = 1
| |
| - | for i = 2:m+1
| |
| - | c{i,j} = trackingMatrix(i-1,:);
| |
| - | end
| |
| - | end
| |
| - | for i = 1
| |
| - | for j = 2:n+1
| |
| - | c{i,j} = inputMatrix(j-1,:);
| |
| - | end
| |
| - | end
| |
| - | for i = 2:m+1
| |
| - | for j = 2:n+1
| |
| - | c{i,j} = processMatrix;
| |
| - | end
| |
| - | end
| |
| - |
| |
| - | %Rules, Acessing, and Changing
| |
| - | for j = 2:n+1
| |
| - | A = c{1,j}(1);
| |
| - | B = c{1,j}(2);
| |
| - |
| |
| - | for i = 2:m+1
| |
| - | mRNA_pA = c{i,1}(1);
| |
| - | mRNA_pB = c{i,1}(2);
| |
| - | sRNA_A = c{i,1}(3);
| |
| - | sRNA_B = c{i,1}(4);
| |
| - | protein_A = c{i,1}(5);
| |
| - | protein_B = c{i,1}(6);
| |
| - |
| |
| - | %Trx_pA
| |
| - | if A == 1
| |
| - | c{i,j}(1) = 1;
| |
| - | end
| |
| - |
| |
| - | %Trx_pB
| |
| - | if B == 1
| |
| - | c{i,j}(2) = 1;
| |
| - | end
| |
| - |
| |
| - | %Trx_sRNA_A
| |
| - | if A == 1
| |
| - | c{i,j}(3) = 1;
| |
| - | end
| |
| - |
| |
| - | %Trx_sRNA_B
| |
| - | if B == 1
| |
| - | c{i,j}(4) = 1;
| |
| - | end
| |
| - |
| |
| - | %Trl_A
| |
| - | if mRNA_pA == 1 && mRNA_pB == 0
| |
| - | c{i,j}(5) = 1;
| |
| - | end
| |
| - |
| |
| - | %Trl_B
| |
| - | if mRNA_pB == 1 && mRNA_pA == 0
| |
| - | c{i,j}(6) = 1;
| |
| - | end
| |
| - |
| |
| - | end
| |
| - | end
| |
| - |
| |
| - | %Display Results (Column by Column)
| |
| - | [nrows,ncols]= size(c);
| |
| - | %Condense Cell and Display
| |
| - | for i = 1:nrows
| |
| - | for j = 1:ncols
| |
| - | string = num2str(c{i,j});
| |
| - | l = length(string);
| |
| - | r = 1;
| |
| - | s = 1;
| |
| - | t = 0;
| |
| - |
| |
| - | while t ~= 1
| |
| - | if r == l
| |
| - | t = 1;
| |
| - | end
| |
| - |
| |
| - | noSpacesString(s) = string(r);
| |
| - | r = r+3;
| |
| - | s = s+1;
| |
| - | end
| |
| - | c{i,j} = noSpacesString;
| |
| - | end
| |
| - | end
| |
| - | c(:,:)
| |
| - |
| |
| - | %Find Steady States and Corresponding Inputs
| |
| - | counter = 1;
| |
| - | for i = 2:m+1
| |
| - | for j = 2:n+1
| |
| - | if c{i,j} == c{i,1}
| |
| - | completeMatrix{counter,1} = c{i,j}; %Steady state values
| |
| - | completeMatrix{counter,2} = num2str([i-1,j-1]); %Location (n x m) within results area
| |
| - | completeMatrix{counter,3} = c{1,j}; %Corresponding input values
| |
| - | counter = counter+1;
| |
| - | end
| |
| - | end
| |
| - | end
| |
| - | SS_mXn_Input = completeMatrix
| |
| - |
| |
| - | time = 0:processNum;
| |
| - | %plot(time,,time,,time,,time,,time,,time,)
| |
| - |
| |
| - | %New Cell for Specific Cases
| |
| - | % d(:,1) = c(:,1);
| |
| - | % for j = 2:n+1
| |
| - | % C1 = '0000';
| |
| - | % C2 = '0011';
| |
| - | % C3 = '0100';
| |
| - | % C4 = '0111';
| |
| - | % C5 = '1000';
| |
| - | % C6 = '1011';
| |
| - | % C7 = '1100';
| |
| - | %
| |
| - | % a = {C1, C2, C3, C4, C5, C6, C7};
| |
| - | % for b = 1:1:length(a)
| |
| - | % if strcmp(c(1,j),a(b)) == 1
| |
| - | % d(:,b+1) = c(:,j);
| |
| - | % end
| |
| - | % end
| |
| - | % end
| |
| - | % d
| |
| - | ##### SOURCE END #####
| |
| - | --></body>
| |
| - | </html>
| |
| | | | |
| | | | |

 "
"