Theory
In this section we will review the theory behind our biological work.
Phototaxis
Bacterial flagellar motility
Bacteria have evolved many modes of propulsion for the microscale environments they inhabit. At these scales materials behave very different than at macroscale, and of particular interest to us is the way liquids seem more viscous [1]. One of the many ways bacteria move around in liquids, is by means of flagella. A single flagellum is a thin filament around 100-150 Å thick, that extends many cell lengths out from the cell [2]. It consists mainly of flagellin subunits that assemble into a helical structure forming a long hollow cylindrical filament [3]. In E. coli the mean number of flagella per cell is 4 [4], but there is a wide variance between strains, and even between individual cells of each strain. The environment around the cell also has a large influence on how many flagellae are present, or if they are present at all [4]. Flagella rotate to generate force that allows bacterial cells to swim through fluids in characteristic patterns, more on which later, in search of better conditions for proliferation or survival [5]. Normally flagellated strains of E. coli can achieve speeds up to 20µm/sec [4], and considering a cell length of only 1-2µm, this is an impressive feat indeed.
A flagellum is anchored to the cell body by a large, wheel-like protein complex spanning both inner and outer membranes [4]. Through this complex subunits are secreted to the tip of the flagellar tube [3], thus elongating the filament (This mechanism is slightly different in archaea, where the filament is assembled from the base of the flagellum). The membrane anchor also functions as a rotary engine, driven by the proton motive force [4]. In E. coli the flagellar motor rotates at up to 300hz and can turn either clockwise or counter-clockwise [4], both resulting in a distinct movement pattern for the cell.
Spinning in the counter-clockwise direction, the flagella will twist into a bundle in the shape of a corkscrew, and create a linear driving force [4], propelling the cell in a straight line through the liquid. This form of movement is termed run. Spun in the clockwise direction one might then expect the cell to reverse, but this is not the case. Instead the flagellar bundle will unwind, thereby creating chaotic movement [4]. This movement reorients the cell randomly and is termed tumbling. A cell will typically run for some time, then change it’s orientation by tumbling, and then run again [5]. The direction of flagella rotation is controlled by the binding of a cytosolic protein CheY, more on which later [4].
Different taxis pathways that steer cells towards favorable conditions and away from danger work by regulating the frequency of tumbling events [5]. An example is when a bacterium gets close to the source of a lethal toxin, then intracellular pathways will increase the frequency of tumbling events, in effect preventing the cell from dying. Since the frequency of tumbling events will decrease if the cell is going in a direction away from the toxin, it will “encourage” the cell to continue in that direction. In the case of an attractant such as an increase in nutrient concentration, the pattern will be opposite, so that the cell is encouraged to continue towards the source of the attractant. This form of movement, combining tumbling and running, with regulation of the tumbling frequency is termed a biased random walk [5].
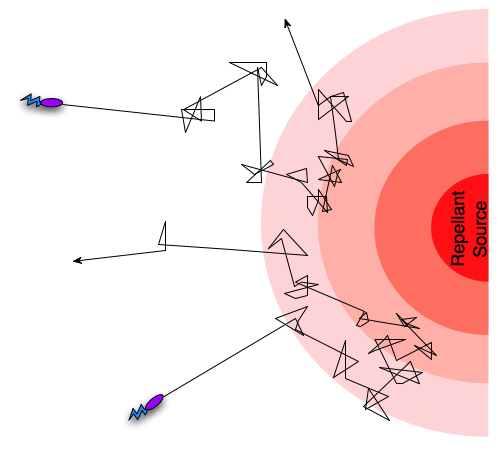
Figure 1: A biased random walk motion pattern.
To understand how this can work we need a simplified understanding of the chemotaxis pathway at a molecular level. Chemotactic receptors can both increase and decrease tumbling frequencies to generate a biased random walk behavior [6]. Increased tumbling is achieved through a phosphorylation cascade beginning with the binding of a repellant to a transmembrane receptor [4]. The receptor is linked to two proteins, CheW and CheA [4]. CheA is a histidine-kinase that will autophosphorylate when the repellant binds [4]. The phosphoryl group is then transferred to CheY [4]. The flagellar motor complex has high affinity for phosphorylated CheY (CheY-p), and binding reverses the mode of movement from run to tumble [7]. CheY-p is continuously dephosphorylated back to CheY by CheZ [8]. A receptor sensing an attractant might instead switch from the default active CheA state to an inactive state when it’s ligand is bound, thus decreasing CheY phosphorylation [6].
Photosensor
In our system we want to be able to control the amount of flow in the channel, through a remote signal. The signal we have chosen is light since we want to avoid altering the chemical composition of the fluid running through the channel. Having looked at previous iGEM work on light sensitive systems which have all been focused on transcriptional regulation (e.g. I15010), we realized that we would need a different approach for the fast response times our system requires. We have therefore focused our work on proteorhodopsins that integrate into the chemotaxis pathway, giving us very fast responses to light stimulation.
Our construct centers around a synthetic protein created by Spudich et al. [9]. It is a fusion chimeric protein that consists of an archaeal proteorhodopsin SopII (Sensory Rhodopsin II) and its transducer protein HtrII, both from Natronomonas pharaonis. These are coupled to a Tar domain from a transmembrane receptor from Salmonella enterica. The Tar domain is the part of the receptor that couples with CheA and CheW, and although it is taken from a different species, it has been shown to work in E. coli as well [10]. In the construct we are working with light acting as an attractant, reducing the tumbling rate upon illumination (see picture). This might help us to control our pumping power, by decreasing the fraction of bacteria tumbling in the channel by increasing light stimulus, thus promoting linear drive. The photosensor should be most active in light with a wavelength of about 500nm, according to the original article. We have used DNA sent to us from the original authors to isolate the coding sequence for the protein generator.
Note that although the bacteria will be stationary in our system, since they are glued to the inner surface of the flowchannel, our construct in reality confers a phototactic ability to E. coli.
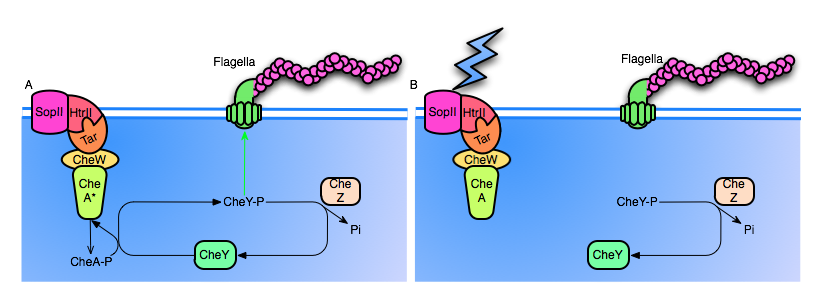
Figure 2: A. The SopII proteorhodopsin has not been activated. Note that CheA is active by default, continuosly autophosphorylating itself, and cycling back to it's unphosphorylated state by transfering the phosphoryl group to CheY. High levels of CheY-p will induce tumbling motion in the flagella. Also, note that CheZ continuously dephosphorylates CheY.
B. SopII is hit with a photon, causing conformational change of the entire complex. The inactivation of CheA halts production of CheY-p, and CheZ rapidly dephosphorylates the remaining CheY-p, resulting in a reduced frequency of tumbling.
BioBrick design

Photosensor coding biobrick: K343003

Photosensor composite part: K343007
Retinal Generator
Retinal requirements of light-sensing proteins
Proteorhodopsins belong to a group of molecules consisting of a protein called an opsin with a cromophore(usually a retinal enantiomer) attached as a prosthetic group. The retinal chromophore is the molecule responsible for the initial light-activation, as it undergoes photoizomerization after being struck by a photon. It is this change in conformation of the retinal molecule that is relayed through the entire rhodopsin-transducer complex to activate/deactivate the CheW/A complex in the cytosol. Thus either an external or an internal supply of retinal, generated by means of genes coding for enzymes in the retinal biosynthesis pathway are required, if we wish to see phototactic behaviour in our cells.
These light-sensitive pigments can be found in a large variety of organisms from archaea and bacteria to both uni- and multicellular eukaryotes. Many plants and microbes have complete retinal biosynthesis pathways in their genomes, to help drive their rhodopsins. In these organisms rhodopsins play an essential role, not only for photosensing but also directly in energy production. In fact in some organisms rhodopsins are used to create proton motive force directly by pumping protons out into the extracellular space using light energy to drive the process. Humans and other animals on the other hand often only have enzymes coding for the final steps of the pathway, more on which later. They rely on a supply of retinal precursors or vitamin A (a group of molecules consisting of retinal and it's metabolites) in their diet. This is why vitamin A deficiency causes night-blindness as an early symptom in humans.
Retinal biosynthesis
Retinal is also synthesized from the enzymatic cleavage of some carotenes. In our system we focus on cleavage of beta-carotene, partly because it yields 2 all-trans retinal molecules which are the molecules we desire, and partly because the beta-carotene biosynthesis pathway has already been introduced to E. coli by the Cambridge 2009 iGEM team.
The Cambridge construct uses genes from the plant pathogen Pantoea ananatis and our construct completes the pathway to retinal with a gene from the common fruit fly, Drosophila melanogaster.
The Cambridge 2009 construct consists of four genes crtE, crtB, crtI and crtY from P. ananatis that together make up the pathway that converts farnesyl pyrophosphate to beta-carotene, which is a precursor for retinal. Farnesyl pyrophosphate is naturally present in E. coli.
• crtE encodes the protein geranyl-geranyl pyrophosphate synthase that converts farnesyl pyrophosphate to geranyl-geranyl pyrophosphate by elongating it by one unit of isopentenyl.
• crtB encodes the protein phytoene synthase and synthesizes phytoene by putting together two molecules of geranyl-geranyl pyrophosphate whilst cutting off 2 molecules of pyrophosphate.
• crtI encodes the protein phytoene dehydrogenase and converts phytoene to lycopene by converting the trans bond to a cis bond and adding more cis double bonds.
• crtY encodes the protein lycopene B-cyclase and converts lycopene to beta-carotene.
The pathway (including the step that generates retinal) is summed up below:
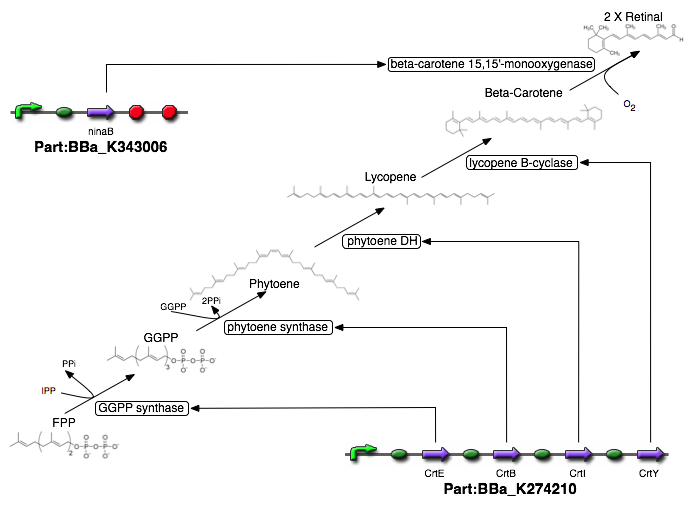
Figure 3: The retinal Biosynthesis pathway.
To introduce the final step from beta-carotene to retinal, we use the gene ninaB from D. melanogaster. This gene encodes the protein beta-carotene 15,15’-monooxygenase, which cleaves beta-carotene to produce two molecules of trans-retinal under the consumption of oxygen.
We have inserted the part K343006 into a different plasmid from the K274210 part since both parts are very long, so a plasmid containing both wouldn't have been viable.
BioBrick design

Retinal coding biobrick: K343001
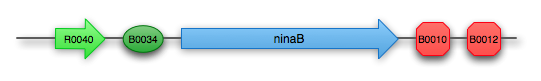
Retinal composite part: K343006
Further use of the retinal BioBrick
Role in light-based signal transduction
Since retinal plays such an essential role in photosensing in both eukaryotes as well as bacteria and archaea, all work with rhodopsins and proteorhodopsins will need a retinal supply to function. This supply might come from the external environment, but it is an appealing thought that we might be able to supply the retinal internally. Our project centers around phototaxis, but constructs combining photorhodopsins with other membrane associated tyrosine kinases may also be imagined, opening vast posibilities for regulation of phopsphorylation cascades using light as input. In such systems, retinal biosynthesis could play a very valuable role.
Hyperflagellation
Background
Motility can be a very beneficial quality for a microorganism. Evolution has therefore provided bacteria with two general means of transportation; the flagella and the pili. These qualities allow the bacteria to move away from a hostile environment and towards more favorable conditions. Flagella and pili are however viewed as a virulence factor as they also serve as an advantage in colonizing a host organism and yet they can cause a strong immune response [11].
Many organisms are able to synthesize a flagellum, if the external environment calls for it. The synthesis of a flagellum is a huge and energy consuming process and is therefore tightly regulated by the bacteria’s external environment. One of the most well characterized flagellation systems is the one found in E. coli. Here at least 50 genes are involved in the hierarchical synthesis and operation of the flagella. These genes are sorted into 15 operons which are expressed in a transcriptional cascade separated into three classes. Class I consists of the master operon flhDC. The active FlhDC protein is a hexamer organized into an FlhD4C2 complex with a computed value of 96,4kDa [12]. The homodimeric FlhC protein is able to bind DNA, while the FlhD homodimers are not. The formation of the FlhDC complex however, stabilizes and increases the DNA binding ability [13]. The transcription of flhDC is heavily regulated by nutritional and environmental conditions. Flagellum synthesis is inhibited at high temperatures, at high salt concentrations, at extreme pH or in the presence of carbohydrates, low molecular alcohols or DNA gyrase inhibitors, as these conditions stimulate growth as opposed to motility [14]. Because the flagellum synthesis is so energy consuming, the process is not started unless the environment calls for motility rather than growth. In fact, in situations where nutrition is plenty over a long period, the bacteria will focus on growth and over time lose the ability to synthesize the flagellum, as seen with the E. coli strain MG1655 localized in mouse intestines [15].
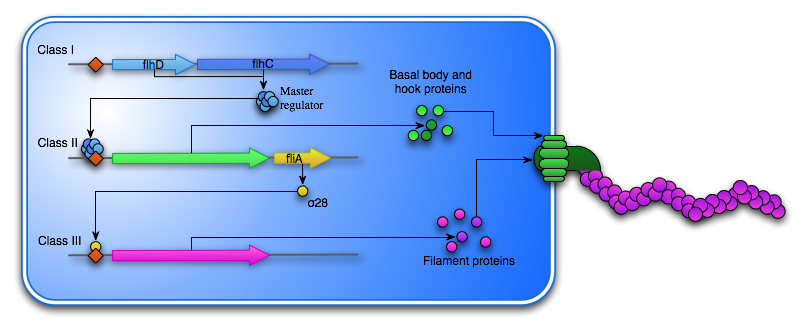
The FlhD4C2 hexamer acts as a transcription factor for the Class II genes, which encodes the basal body, that is embedded in the cell membrane as well as hook proteins, which are transported to the cell exterior through the basal body. Another Class II gene is the σ28 transcription factor, which is responsible for the transcription of the Class III genes. This includes fliC, which encodes the flagellin subunit that composes the flagella “tail”. To ensure that the Class III genes are not transcribed before the assembly of the basal body and the hook is complete another Class II protein FliM acts as an anti-sigma factor and bind σ28, thereby preventing the transcription of fliC.
Several studies regarding the motility of E. coli has shown the expression of the flhDC operon to be crucial [15][16]. These focused on insertion sequence (IS) elements upstream of the flhDC regulon. IS are sequences that can be inserted randomly within the DNA and therefore serve as an important factor in the plasticity of the E. coli genome as well as in many other organisms. Generally they do not encode any genes apart from those responsible for their movement within the genome, however, they can also serve as activators of neighboring genes, by disrupting repression or by the formation of hybrid promoters [16]. In the beforementioned studies, bacteria containing an activating IS upstrem of the flhDC operon showed an increased motility compared to bacteria without this IS. It is therefore resonable to asume that by placing a constitutive active promoter in front of the flhDC operon, hyperflagellation will be induced.
Our system
As we are trying to create a microflow in our system using flagella, we want to increase the flow created by a single bacteria by increasing the number of flagella on the bacteria. However, inducing flagellation in a wildtype E. coli strain requires too much of the bacterial environment. In order to avoid the tight control of the flhDC operon, we insert it into a plasmid backbone containing a constitutive active promoter. As we bypass the original regulations and continuously express the master regulon of the flagellum cascade we hope to see a significant difference in motility of the cells containing our composite part.
As E. coli already express the flhDC operon, we isolate this coding sequence by purifying genomic DNA from the E. coli MG1655 strain and then amplifying the operon using PCR with specially designed primers, before assembly and the insertion into the plasmid backbone
Furthermore, it will be interesting to see whether the overexpression of the flhDC operon will have an effect on the bacterial growth.
Biobrick design
FlhD,C mutated coding sequence: K343000
FlhDC composite part: K343004
References
[1] Li T-D, Gao J, Szoszkiewicz R, Landman U, Riedo E, Structured and viscous water in subnanometer gaps,Phys. Rev. B 75, 115415 (2007)
[2] Samatey FA, et. al.,Structure of the bacterial flagellar protofilament and implications for a switch for supercoilingNature 410, 331-337 (15 March 2001)
[3] Macnab RM, How bacteria assemble flagella Annual Review of Microbiology Vol. 57: 77-100 (October 2003)
[4] Berg HC, The rotary motor of bacterial flagella Annual Review of Biochemistry Vol. 72: 19-54 (July 2003)
[5] Berg HC, Chemotaxis in bacteria Annu Rev Biophys Bioeng. 1975;4(00):119-36.
[6] Zhao J, Parkinson JS, Mutational Analysis of the Chemoreceptor-Coupling Domain of the Escherichia coli Chemotaxis Signaling Kinase CheA , Journal of Bacteriology, May 2006, p. 3299-3307, Vol. 188, No. 9
[7] Sarkar MK, Paul K, Blair D,Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli, Published online before print May 3, 2010, doi: 10.1073/pnas.1000935107
[8] Hess JF, Oosawa K, Kaplan N, Simon MI, Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis., Cell. 1988 Apr 8;53(1):79-87.
[9] Trivedi VD, Spudich JL, Photostimulation of a sensory rhodopsin II/HtrII/Tsr fusion chimera activates CheA-autophosphorylation and CheY-phosphotransfer in vitro., Biochemistry. 2003 Dec 2;42(47):13887-92.
[10] Jung K-H, Spudich EN, Trivedi VD, Spudich JL, An Archaeal Photosignal-Transducing Module Mediates Phototaxis in Escherichia coli, Journal of Bacteriology, November 2001, p. 6365-6371, Vol. 183, No. 21
[11] Josenhans, C. and Suerbaum, S. (2002) The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291, 605-614
[12] Wang, S. et al. (2006) Structure of the Escherichia coli FlhDC Complex, a Prokaryotic Heteromeric Regulator of Transcription. Journ. of mol. Biol. 355, 4, 798-808
[13] Claret, L. and Hughes, C. (2000) Functions of the Subunits in the FlhD2C2 Transcriptional Master Regulator of Bacterial Flagellum Biogenesis and Swarming. J. Mol. Biol. 303, 467-478.
[14] Li, C., Louise, C.J., Shi, W. and Adler, J. (1993) Adverse Conditions Which Cause Lack of Flagella in Escherichia coli. J. Bacteriol. 175,
2229-2235.
[15] Gauger, E.J. et al (2007) Role of Motility and the flhDC Operon in Escherichia coli MG1655 Colonization of the Mouse Intestine. Infection and Immunity. vol. 75, No. 7. p3315–3324
[16] Baker, C. S. et al (2004) Increased Motility of Escherichia coli by Insertion Sequence Element Integration into the Regulatory Region of the flhD Operon Journal of Bacteriology, Vol 186. No22. p7529–7537
 "
"









