Team:Newcastle/End of crack & signalling system
From 2010.igem.org
RachelBoyd (Talk | contribs) (→Expected results) |
Swoodhouse (Talk | contribs) (→Testing and Characterisation) |
||
| Line 67: | Line 67: | ||
===Testing and Characterisation=== | ===Testing and Characterisation=== | ||
| - | + | ====Selection for integration==== | |
To select for integration of the plasmid into the chromosome, ''B. subtilis'' will be tested for the ability to hydrolyse starch. Integration of the BioBrick will be done by homologous recombination at the amyE gene, therefore destroying endogenous expression of amylase. Colonies that are not able to break down starch on agar plate will be selected and cultured for further test. Colonies that do not contain the integrated BioBrick will be able to hydrolyse starch, therefore forming a white halo around the colony as iodine interacts with starch to form blue colour. | To select for integration of the plasmid into the chromosome, ''B. subtilis'' will be tested for the ability to hydrolyse starch. Integration of the BioBrick will be done by homologous recombination at the amyE gene, therefore destroying endogenous expression of amylase. Colonies that are not able to break down starch on agar plate will be selected and cultured for further test. Colonies that do not contain the integrated BioBrick will be able to hydrolyse starch, therefore forming a white halo around the colony as iodine interacts with starch to form blue colour. | ||
| - | Isolation of subtilin from ''B. subtilis'' ATCC6633 | + | ====Isolation of subtilin from ''B. subtilis'' ATCC6633==== |
1. ''B. subtilis'' ATCC6633 were grown in 250 ml flasks containing 100 ml of BHI broth and incubate for 48 h at 32°C in shaker at 125 cycles/min. | 1. ''B. subtilis'' ATCC6633 were grown in 250 ml flasks containing 100 ml of BHI broth and incubate for 48 h at 32°C in shaker at 125 cycles/min. | ||
Revision as of 15:41, 13 August 2010

| |||||||||||||
| |||||||||||||
Contents |
End of crack cell-signalling system
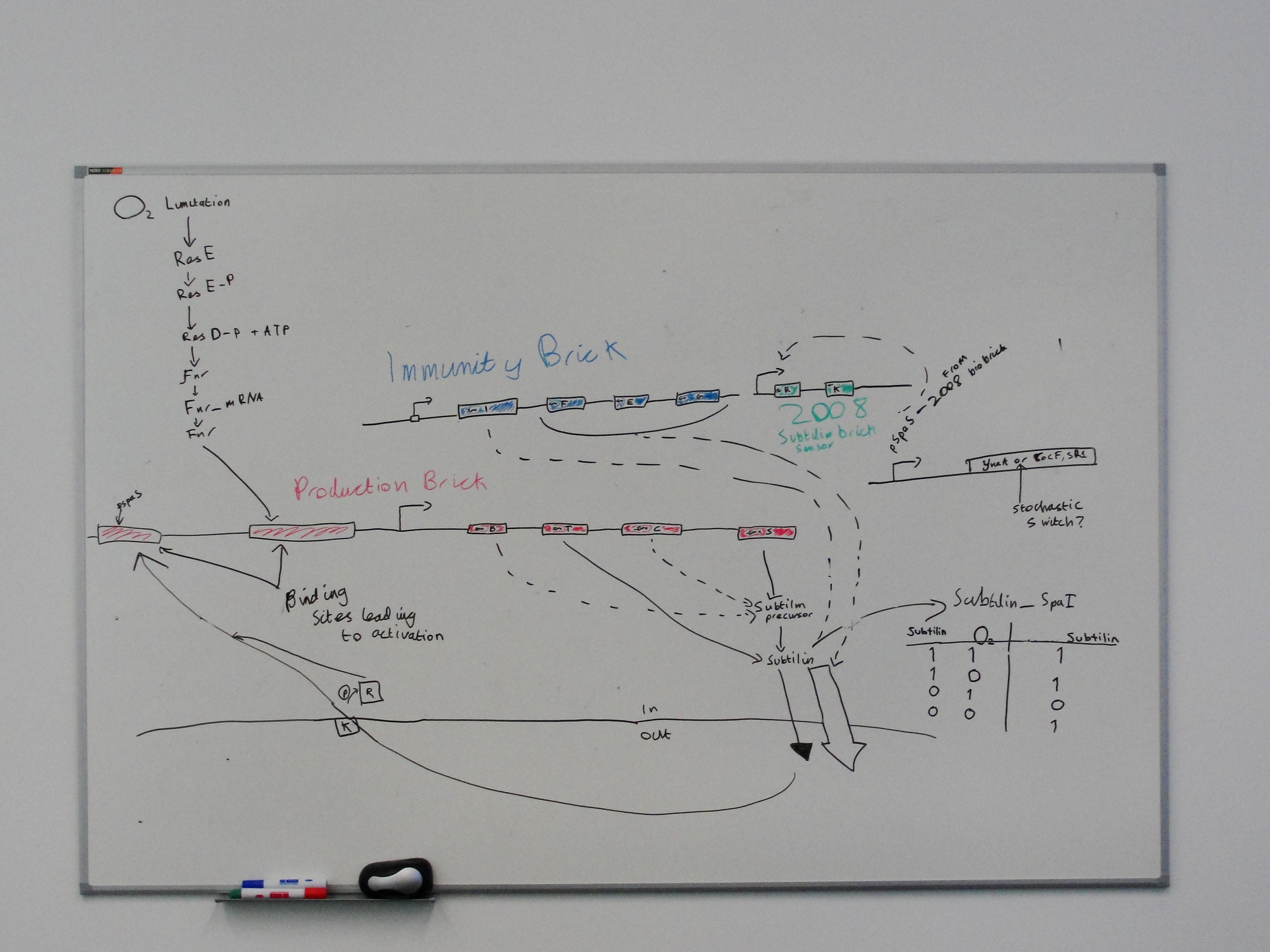
We aim to use a subtilin-based cell signalling system to trigger calcium carbonate precipitation and filament formation once our bacteria have reached a sufficient density inside a microcrack.
Subtilin is a lantibiotic peptide produced naturally by Bacillus subtilis ATCC 6633 but not by Bacillus subtilis 168. We aim to produce two BioBricks: one for production of subtilin, and, since subtilin is a lantibiotic, one for subtilin immunity. A subtilin-sensing BioBrick was produced by Newcastle's 2008 iGEM team.
The spaIFEG gene cluster encodes two separate subtilin immunity systems, both present in ATCC 6633. Together they provide a high degree of subtilin resistance.
Research
2008 Subtilin-sensing BioBrick
- A type 1 antimicrobial peptide [AMP] or lantibiotic produced by B. subtilis
- NB. Lantibiotic = peptide-derived antibiotics with high antimicrobial activity against various Gram-positive bacteria, including pathogenic bacteria such as propionibacteria, staphylococci, clostridia, enterococci and streptococci.
- NB. Subtilin not same as Subtilisin!!!!!
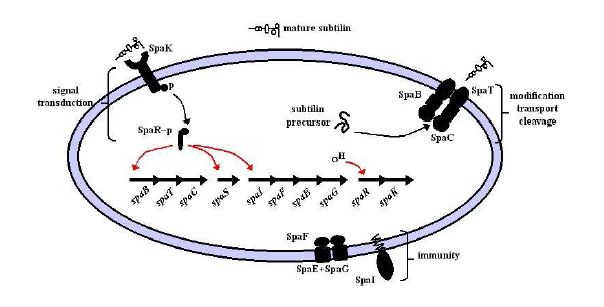
- Production of subtilin involves the spa gene cluster encompassing 10 genes, spaBTCSIFEGRK
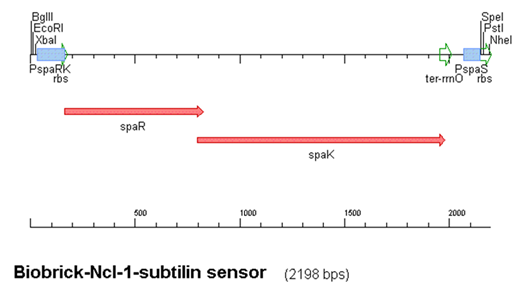
The construct above contains:
- spaRK promotor
- spaR (subtilin peptide antibiotic Regulation) - the 220 amino acid product of this gene usually regulates the downstream production of subtilin antibiotic. It has an N-terminal domain that can be phosphorylated and a C-terminal domian that has DNA binding properties [1]
- spaK (subtilin peptide antibiotic Kinase) - this gene codes for a 325 amino acid histadine kinase peptide that phosphorylates the N-terminus of SpaR [2]. This activates the DNA binding ability of the C-terminus of SpaR, which in turn initiates transcription of the downstream gene. In the case of our construct, this gene is gfp.
- rrnO transcriptional terminator
- spaS promotor - a strong promotor inducible by upstream activation of spaRK. It can be placed in front any gene to regulate its activity.
Production and Immunity Bricks
- spaBTCS genes are required for production.
- spaIFEG genes are required for immunity.
- spaB and C are modifiers of the subtilin precursor.
- spaT is a transporter.
- spaS is the gene coding for the subtilin precursor peptide.
- spaI 's gene product sequesters subtilin.
- spaFEG genes code for transporters.
Image from Modeling Subtilin production in Bacillus subtilis Using Stochastic Hybrid Systems. Hu et al.
Coding Sequence
Cloning Strategy
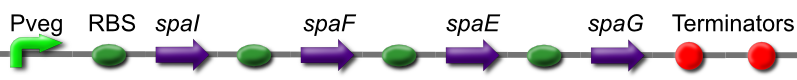
Subtilin Immunity BioBrick
The spaIFEG gene cluster encodes two separate subtilin immunity systems, both present in ATCC 6633. Together they provide a high degree of subtilin resistance.
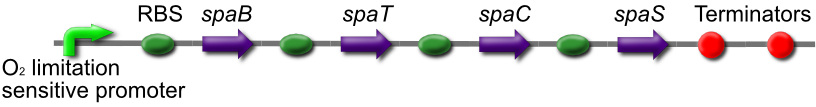
Subtilin production BioBrick
Biochemical pathway
Computational Model
Characterisation
Testing and Characterisation
Selection for integration
To select for integration of the plasmid into the chromosome, B. subtilis will be tested for the ability to hydrolyse starch. Integration of the BioBrick will be done by homologous recombination at the amyE gene, therefore destroying endogenous expression of amylase. Colonies that are not able to break down starch on agar plate will be selected and cultured for further test. Colonies that do not contain the integrated BioBrick will be able to hydrolyse starch, therefore forming a white halo around the colony as iodine interacts with starch to form blue colour.
Isolation of subtilin from B. subtilis ATCC6633
1. B. subtilis ATCC6633 were grown in 250 ml flasks containing 100 ml of BHI broth and incubate for 48 h at 32°C in shaker at 125 cycles/min. 2. Culture media were centrifuged at 10000g for 15 min and the supernatants were filtered through 0.22 m membrane. 3. The pH of the filtrates has to be between 7.0 to 8.0 pH. 4. Store the filtrates at -20°C for further use.
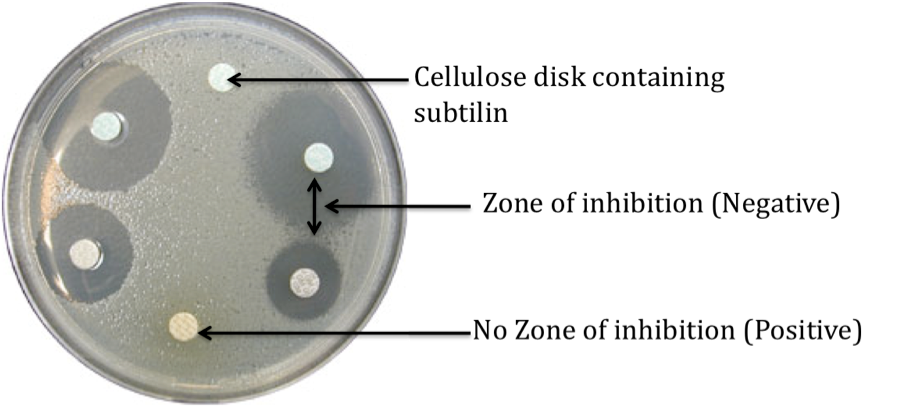
Characterisation of BioBrick
1. Make up 4 LB agar plates and inoculate with the following organism: 1. Negative Control – B. subtilis 168 2. Positive Control – B. subtilis ATCC6633 3. Test (Duplicates) – B. subtilis 168 containing subtilin immunity BioBrick 2. Apply 20 l of the cell free filtrate containing subtilin onto cellulose disks (6 mm) and place onto the LB agar plates. 3. Plates were incubated for 24 h at 37 °C. 4. Zone of inhibition were then measured.
Expected results
| 1. Negative control – Zone of inhibition to be observed |
| 2. Positive control – Zone of inhibition not observed |
| 3. Test (Duplicates) – Zone of inhibition not observed or a smaller zone of inhibition as compared to the negative control |
References
A.S. Motta and A. Brandelli. (2002) Characterization of an antibacterial peptide produced by Brevibacterium linens. Journal of Applied Microbiology 92, 63-70.
A.S. Motta, F. Cladera-Olivera and A. Brandelli (2004) Screen for antimicrobial activity among bacteria isolated from the amazon basin. Brazilian Journal of Microbiology 35, 307-310.
 
|
 "
"